IMGT gene name nomenclature for IG and TR of human and other vertebrates IMGT nomenclature for immunoglobulin (IG) and T cell receptor (TR) gene names of human and other jawed vertebrates is based on the 'CLASSIFICATION' concept of the IMGT-ONTOLOGY [1], and follows as closely as possible the Human Gene Mapping Nomenclature rules. Marie-Paule Lefranc, IMGT founder, has been applying this ontology as early as 1988 for the human IGL and IGH loci [2,3], and 1989 for all the genes of the human TRG locus [4-6]:
IMGT gene names and IMGT gene definitions for the human IG [6-10] and TR genes [11,12] have been approved by HGNC, the HUman Genome Organization (HUGO) Gene Nomenclature Committee in 1999.
Note that, in the HUGO symbols, slashes and parentheses are omitted, and capital letters replace the lower-case letters found in some provisional IMGT gene names. Otherwise the gene symbols and all the full names (including slashes and parentheses) are identical in IMGT and HUGO nomenclatures.
IG nomenclature schematic representation
Download as PDFTR nomenclature schematic representation
Download as PDFGeneral rules
Gene names are in capital letters, no Greek letters, no commas, no dots (hyphens are accepted, as for the HLA and MHC genes). IG and TR genes and alleles are not italicized in publications.
Gene names are defined per species and the same gene symbol can be used across different species without any homology evidence between the two described genes. Names of the species are those defined in NCBI taxonomy database (https://www.ncbi.nlm.nih.gov/taxonomy) [13,14]
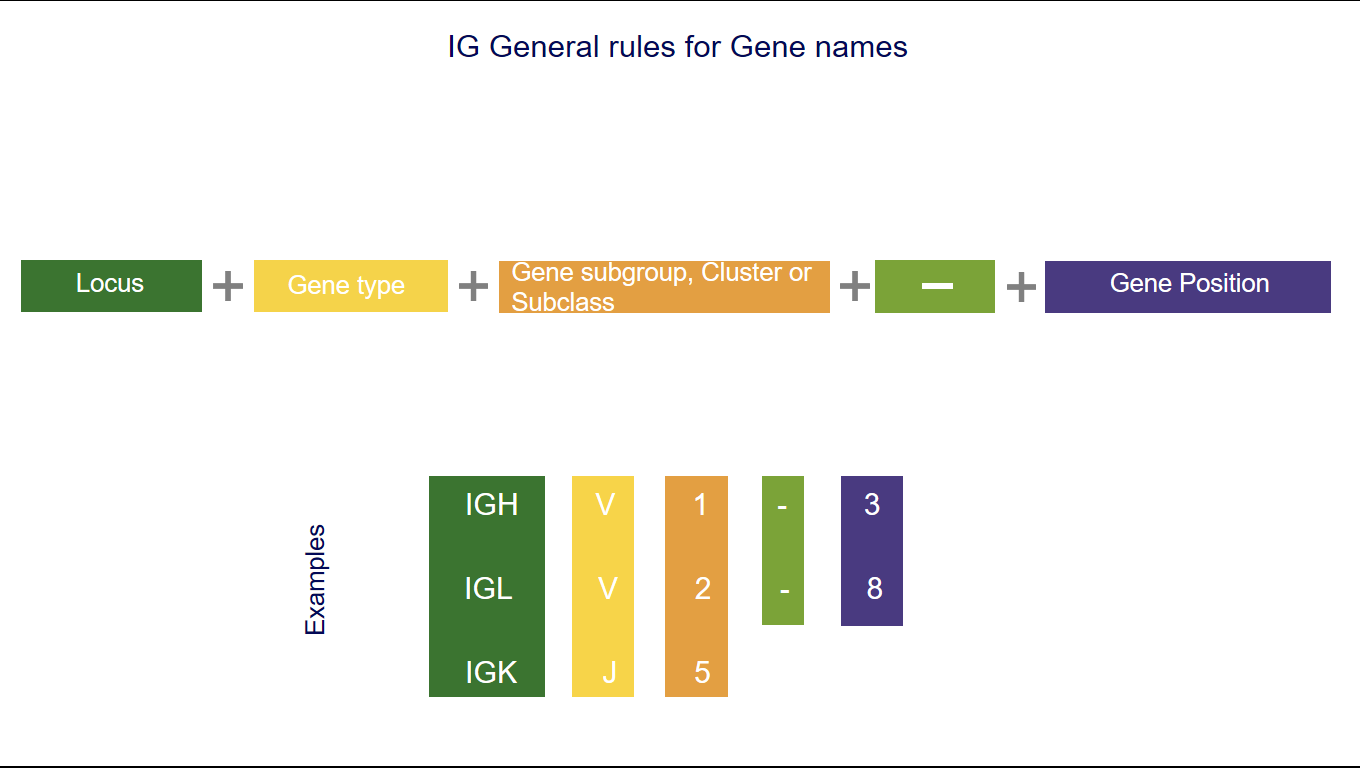
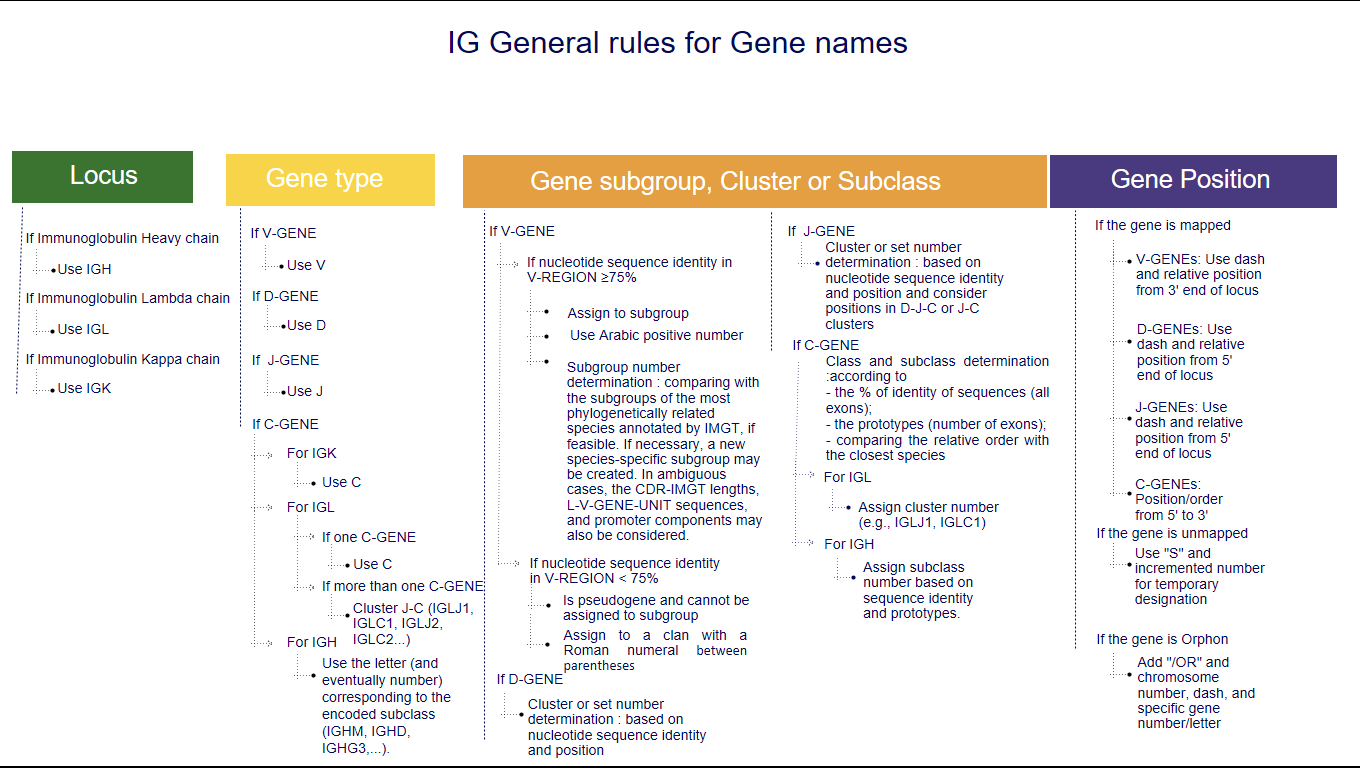
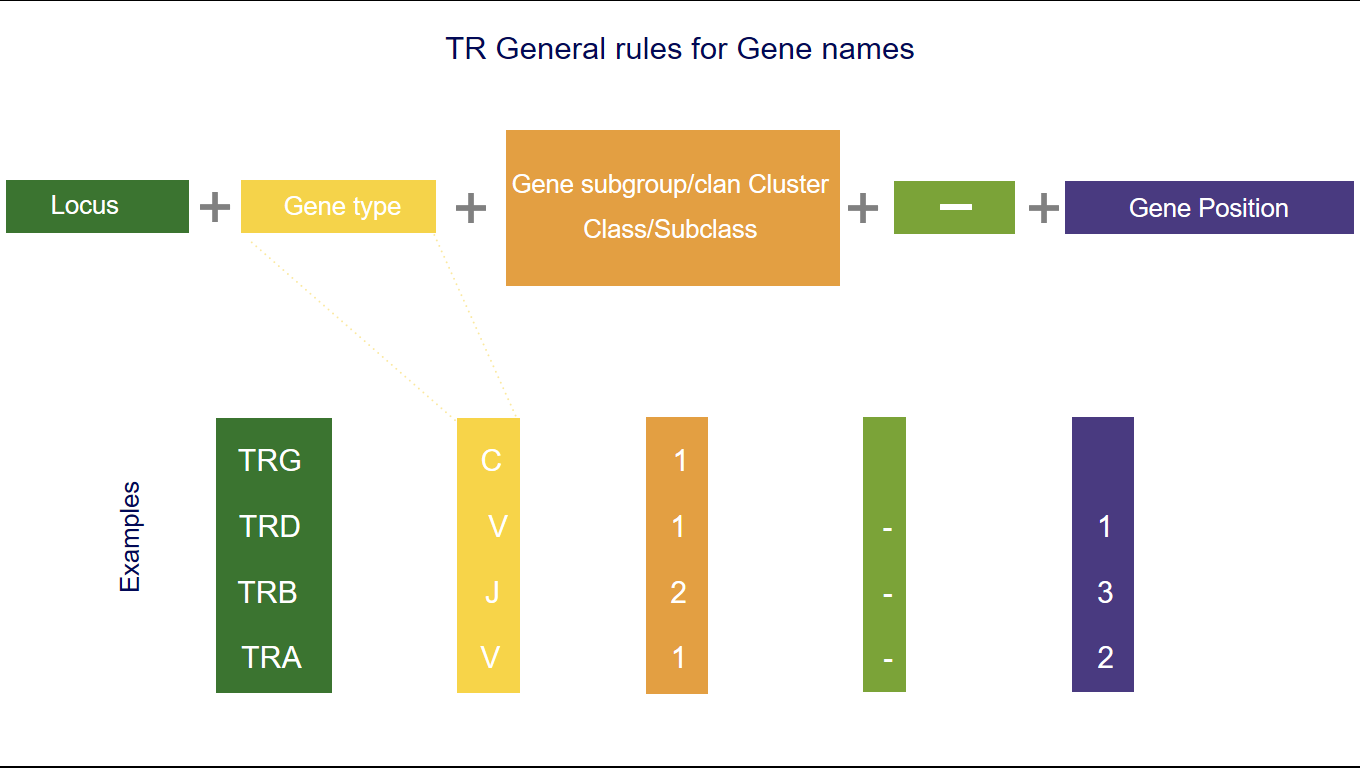
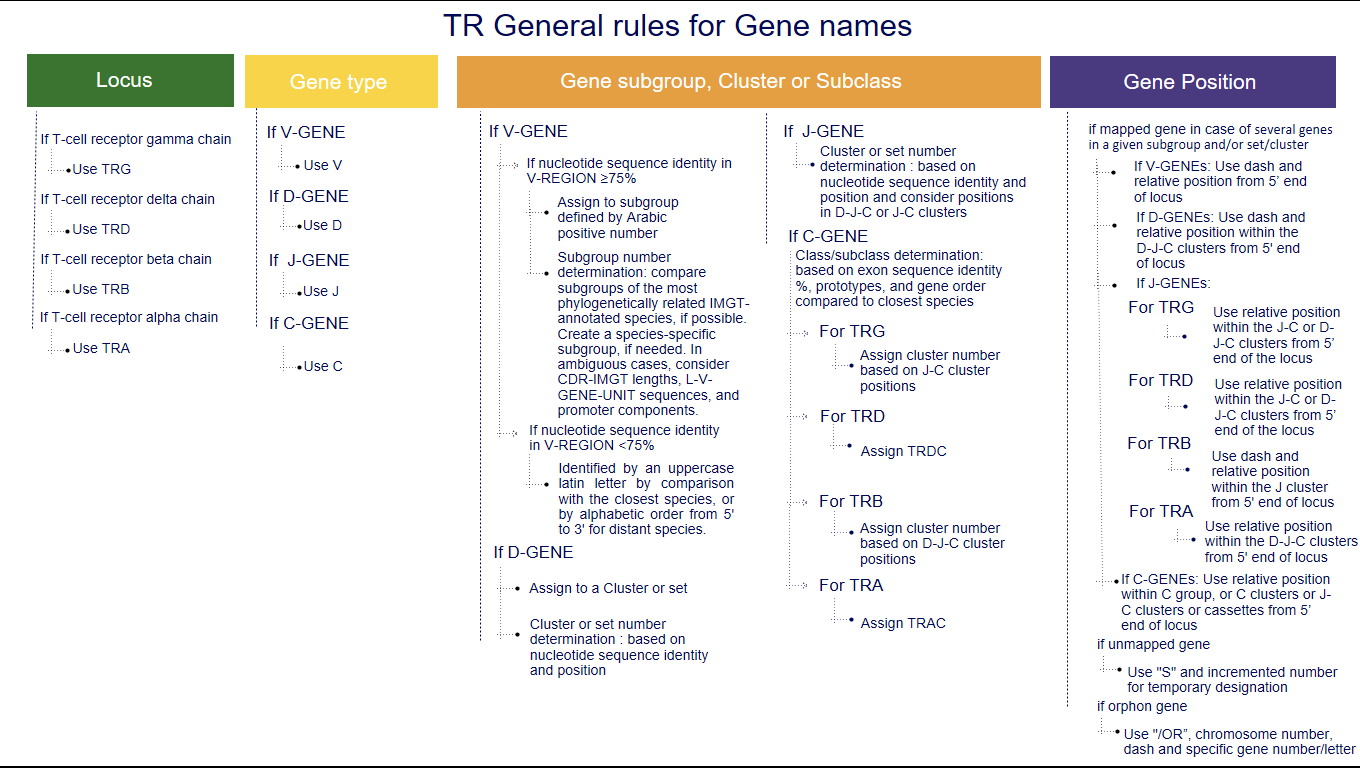
1. The three first letters of a gene name indicate the locus
IGH, IGK, IGL, IGI, TRA, TRB, TRG, TRD [1-5].2. The fourth letter indicates the Gene type
- V (for a variable gene)
- D (for a diversity gene)
- J (for a joining gene)
- C (for a constant gene)
However, for the IGH locus, the constant genes are designated by the letter (and eventually number) corresponding to the encoded class (section 3.3.2) (IGHM, IGHD, IGHG3,...).
3. Following characters correspond to
- the subgroup number or the clan name for V,
- the number of the cluster or sets for D or J,
- the number of the cluster or subclass number of C.
3.1 Subgroup number or clan name for V genes
3.1.1 V genes that can be assigned to a subgroup
Variable genes (whatever their functionality: functional (F), open reading frame (ORF) or pseudogene (P)) of a given species are assigned to the same subgroup if they share at least 75% nucleotide sequence identity in their V-REGION. The subgroup is defined by an Arabic number.The subgroup number is determined by comparison with the subgroups of the most phylogenetically closely related species annotated by IMGT. However, a new subgroup specific to the species may be created.
The CDR-IMGT lengths, the sequences of the L-V-GENE-UNIT and the promoter components may be taken into account to determine the subgroup number in ambiguous cases.
Ex: Homo sapiens IGHV2 , Homo sapiens TRBV13.1.2 V genes that cannot be assigned to a subgroup
Pseudogenes that cannot be assigned to a subgroup due to low % of identity of the V-REGION (degenerated and/or truncated pseudogenes):- are assigned to a Clan for IG. The name of the clan is indicated with a Roman number between parentheses.
- are identified by an uppercase latin letter for TR by comparison with the closest species, or by alphabetic order from 5' to 3' for distant species.
3.2 The number of the cluster or set for a D gene or a J gene
3.2.1 The number of the cluster or set for a D gene
The sets, whenever possible, are defined by comparison with the closest species sets, regarding the identity % of the nucleotide sequence of the D-GENE-UNIT for IGH locus, or their positions in the D-J-C clusters, for the other loci. Clusters are defined by an Arabic number from 5’ to 3’.
3.2.2 The number of the cluster or set for a J gene
The sets, whenever possible, are defined by comparison with the closest species sets, regarding the identity % of the nucleotide sequence of the J-REGION and their positions in the D-J-C-CLUSTER, or J-C-CLUSTER.3.3 The number of the cluster or class and subclass of a constant gene
3.3.1 Cluster for the IGL, TRG and TRB loci
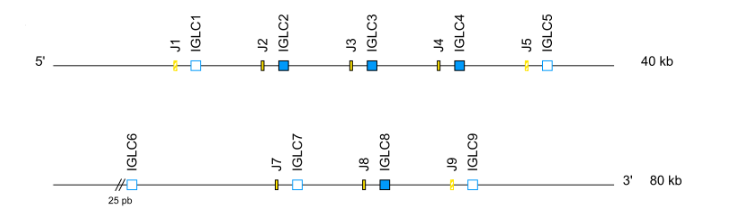
- IGL: cluster J-C (IGLJ1, IGLC1, IGLJ2, IGLC2….) when there is more than one C-GENE
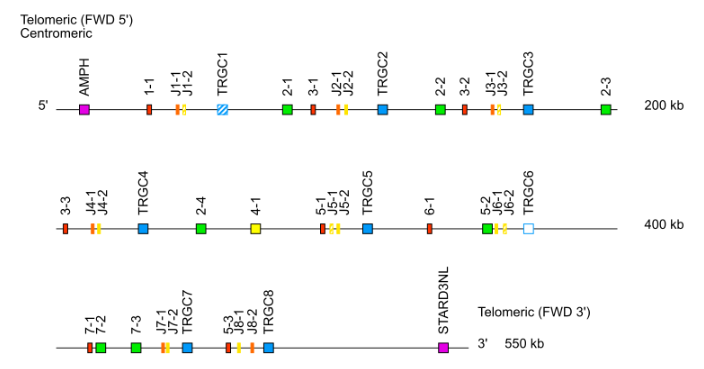
- TRG: cluster J-C (TRGJ1-1, TRGJ1-2, TRGC1, TRGJ2-1, TRGJ2-2, TRGC2…)
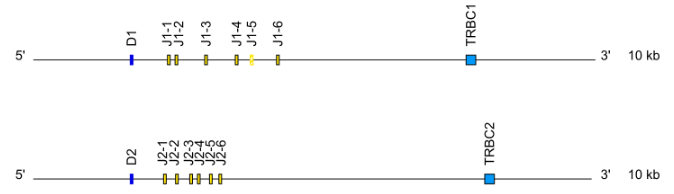
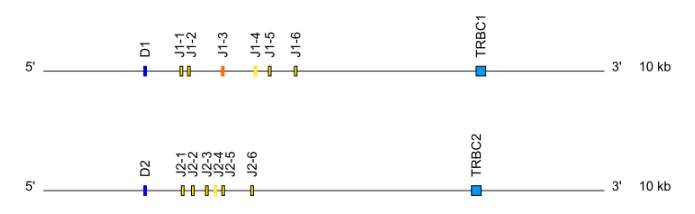
Locus representation: Dog (Canis lupus familiaris) TRG ( V-J-C-CLUSTER). - TRB: cluster D-J-C ( TRBD1,TRBJ1-1 to TRBJ1-n,TRBC1, TRBD2, TRBJ2-1 to TRBJ2-n, TRBC2…)
3.3.2 Class and subclass for the IGH locus
The constant genes are designated by the letter (and eventually number) corresponding to the encoded class and subclass (IGHM, IGHD, IGHG3,...).Determination of the class and subclass of the IG constant gene, according to :
- the % of identity of the sequences (all exons);
- the prototypes (number of exons);
- the comparison with the relative order of the closest species
If more than one gene shares the same subclass, letters in alphabetic order from 5’ to 3’ are appended to the name.
Ex: Gorilla gorilla gorilla IGHG3A, IGHG3B, IGHG3C4. The last part of the gene name indicates the relative position of the gene within the locus and/or in the cluster
- Mapped genes
- Unmapped genes
- Orphon genes
4.1 Mapped genes
4.1.1 Case of IG
- For V genes : A dash (“-”) plus the relative position of the gene within the V cluster, starting from the 3' end of the locus.
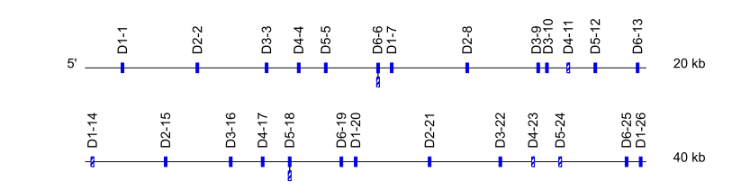
- For D genes : If the D set/cluster was defined as in section 3.2.1, this D set/cluster number is followed by a dash (“-”) plus the relative position of the gene within the D cluster, starting from the 5' end of the locus.
- For J genes : The J set/cluster was defined as in section 3.2.2: For a locus with more than one cluster : dash ("-") plus the relative position of the gene within the J cluster, starting from the 5' end of the locus. For a locus with only one cluster, the position/order in the cluster is given from 5' to 3'. For a locus with many genes in the same set: dash ("-") plus the relative position of the gene within the set, ex IGHJ6 set in the horse.
- For C genes :
- For IGL, the position/order is given from 5' to 3' (ex. Homo sapiens IGLC1, IGLC2).
- For IGK, the position is not indicated, as it is a locus with only one constant gene (ex. Homo sapiens IGKC).
- For IGH, see section 3.3.2.
Ex : Homo sapiens IGHV1-68
4.1.2 Case of TR Only applicable if several genes in a given subgroup and/or set/cluster
- For V genes : a dash (“-”) plus the relative position of the gene within a given subgroup starting from the 5' end of the locus. Ex : Mus musculus TRAV3-1
- For D genes: relative position of the gene within the D-J-C clusters starts from the 5’ end of the locus. Ex : Mus musculus TRBD1
- For J genes :
- For TRG and TRD: Relative position of the gene within the J-C or D-J-C clusters starts from the 5’ end of the locus.
- For TRB: A dash (“-”) plus the relative position of the gene within the J cluster starting from the 5' end of the locus.
- For TRA: Relative position of the gene within the D-J-C clusters starts from the 5’ end of the locus.
- For C genes : relative position of the gene within the C group, or within the C clusters or J-C clusters or cassettes starts from the 5’ end of the locus. Ex : Mus musculus TRBC2
4.2 Unmapped genes
For unmapped genes that have not yet been localized, or are strongly suspected not to be in the right position, a temporary designation is indicated by the letter S for subgroup or sequential, respectively, followed by the incremented number of the last unmapped gene in the subgroup or cluster.
Remark: when an allele named with temporary designation is 100% identical to a newly identified mapped gene, its name is updated with the mapped one and the sequence becomes a sequence from the literature of the mapped gene.
4.3 Orphon genes
Orphon genes are IG and TR genes localized outside of the main locus. Following the same rules for subgroup/set/cluster (if known), "/OR" (for orphon) is added to the name, then the chromosome number (if known), a dash (“-”) and a gene number and/or letter.
Ex: Homo sapiens IGKV2/OR2-1, IGKV1/OR-1, TRBV22/OR9-2.
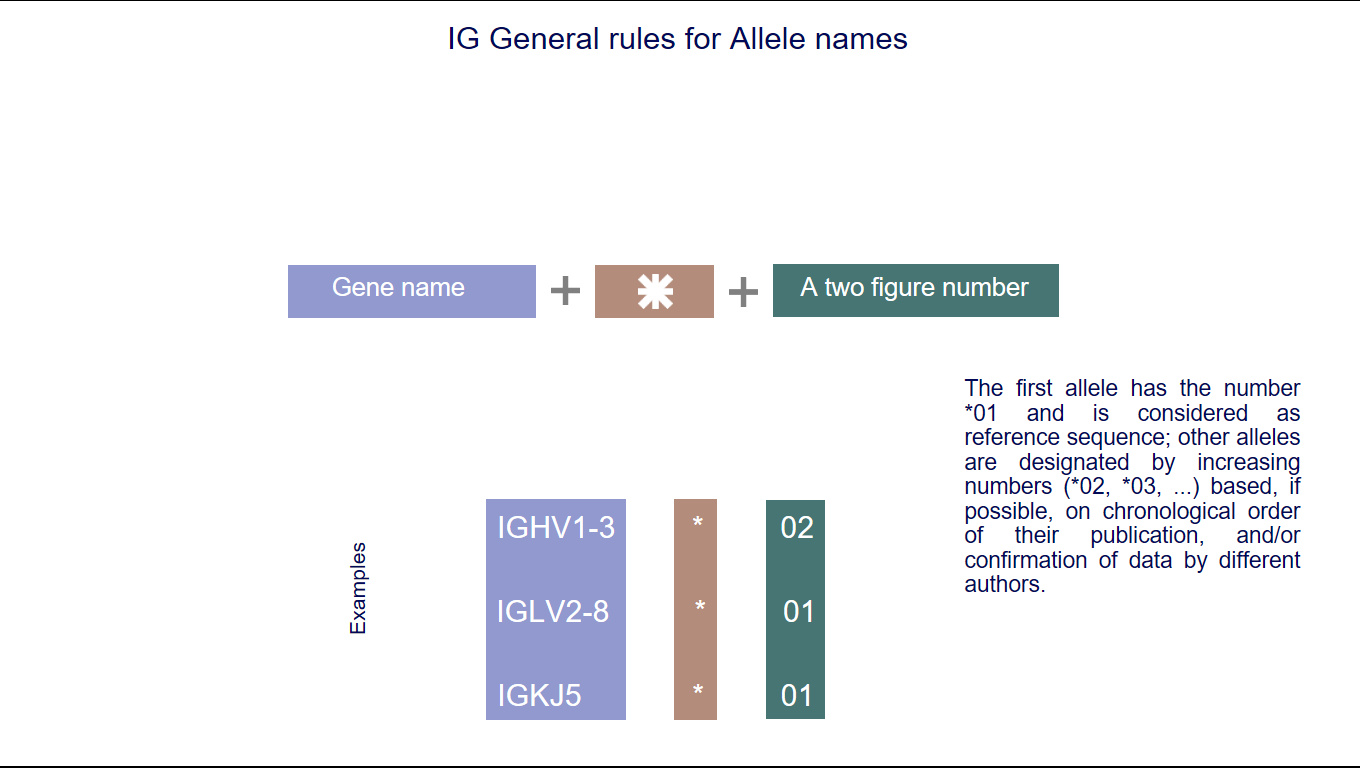
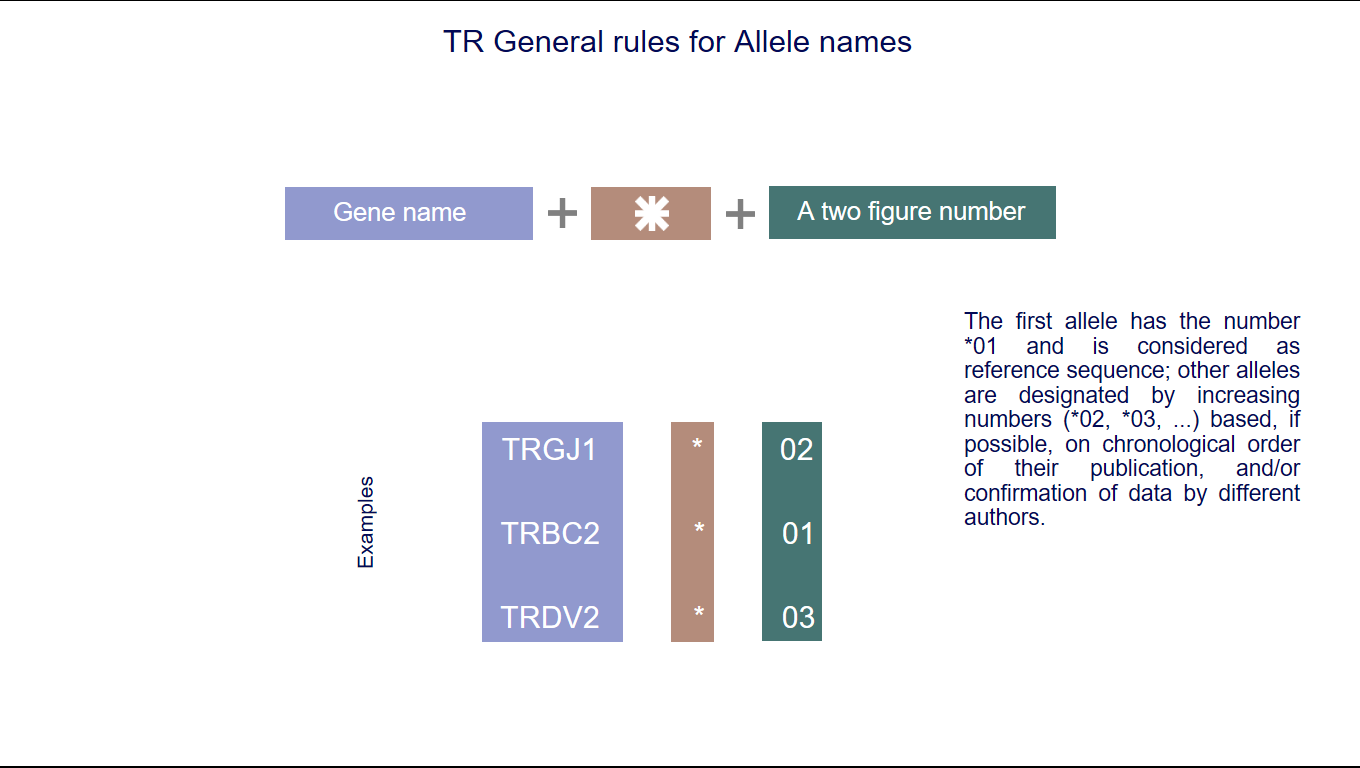
5. IMGT allele names
IMGT allele names comprise the IMGT gene name followed by an asterisk and a two-figure number.
The identification of IG and TR alleles is based on the sequence of the V-REGION, D-REGION, J-REGION and C-REGION. The first allele has the number *01 and is considered as reference sequence; other alleles are designated by increasing numbers (*02, *03, ...) based, if possible, on chronological order of their publication, and/or confirmation of data by different authors. (IMGT allele polymorphism)
Ex : Homo sapiens IGHV1-2*01
There is only one reference sequence per allele. The IMGT Reference sequence should be the first published sequence that meets the following criteria:
- in germline configuration or, for C genes, undefined configuration
- Complete sequence (X-GENE-UNIT)
- Mapped
- Annotated by IMGT
Particular cases
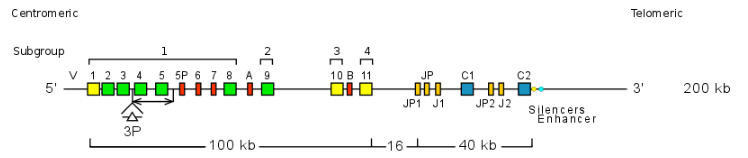
Case 1: Homo sapiens TRGV genes
Homo sapiens TRG locus contains 14 TRGV genes belonging to six subgroups. Gene names do not include the name of the subgroup. This nomenclature is only used for human phylogenetically closely related species annotations.
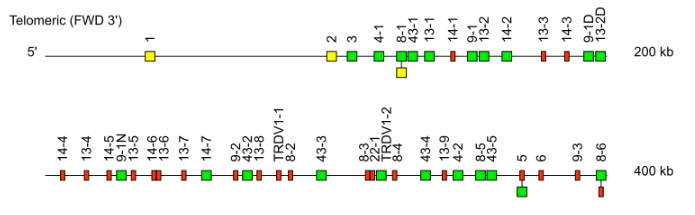
Case 2: Mus musculus IGHV genes
Regarding the Mus musculus IGHV genes, the last part of the gene name indicates the relative position of the gene within a given V subgroup starting from the 3’ end of the locus.
Case 3: TRAV genes which rearrange with TRAJ and TRDJ genes
If a TRAV gene is known to rearrange with TRAJ and TRDD-TRDJ genes, a slash (“/”) is added after the gene name, followed by the TRDV gene without the “TR” prefix.
Ex : Homo sapiens TRAV14/DV4*01
Case 4: Duplicated and triplicated genes
- For individual duplicated and triplicated genes
- For duplicated or triplicated clusters
- For duplicated locus
Case 4A: Individual duplicated and triplicated genes
Duplicated/triplicated genes are genes which share 100% of identity in the V-REGION with an already characterized gene in a different position in the locus.
The gene name is the one assigned to the "initial" gene plus:
- the letter "D" for duplicated gene
- the letter "N" for triplicated gene
Ex: Ovis aries TRAV9-1, TRAV9-1D, TRAV9-1N
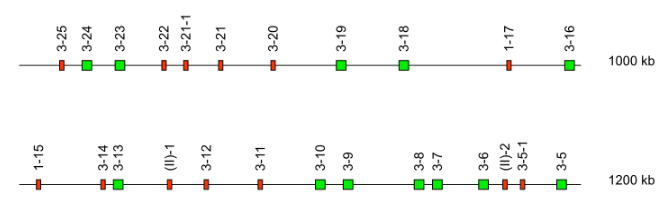
Case 4B: For duplicated or triplicated clusters
Duplicated/triplicated clusters are a group of genes which share a high % of identity in the V-REGION with an already characterized group of genes in a different position in the locus. Additionally, they are characterized by:
- one or more genes share 100% of identity in both clusters
- both clusters show the same sequential order of the subgroups/clans
- There is a equivalent of the distance between the neighboring genes in both clusters
- The gene name follows the rule (case 4A).
Case 4C: For duplicated locus
For duplicated locus (Ex: Homo sapiens IGK), the gene name is the one assigned to the "initial" gene plus the letter "D" for the genes of the duplicated locus, after the number of the subgroup and before the relative position and the dash if any.
Ex: Homo sapiens IGKV3D-25*01
Case 5: Gene insertion(s) and deletion(s)
Case 5A: Gene insertion(s)
Gene insertions and deletions in a locus are determined by comparison with an already annotated reference locus in a given species.
Common genes between the reference locus and the new one are identified: they are named according to the reference locus.
The inserted genes are named according to the classical procedure and to the following rules:
- the inserted gene is named by its subgroup followed by the position of the closest conserved gene in 5’ for IG loci, and in 3’ for the TR loci, a sub position is created by a second addition of a “-” and a number: it will be incremented in case of future new discovered genes, no more than 2 dashes are accepted in IMGT gene names for V gene.
- the latin alphabet letters, from 5' to 3' are added for J gene and C gene .
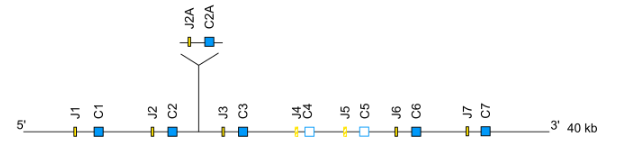
Case 5B: Gene deletion(s)
For missing genes in the locus to be annotated by comparison with the reference locus, their name cannot be used. it is treated as a CNV by deletion.
Case 6: Mus musculus nomenclature
In order to establish the nomenclature of the new Mus musculus strains for instance, the existing rules and nomenclature were taken into account:
- The similarities of the new strains' genes with the ones from IMGT annotated strains (C57BL/6J and 129/Sv, the IMGT reference repertoire) were sought.
- The genes that get 100% similarity with an existing IMGT annotated gene (the IMGT reference repertoire) serve as “anchors”, allowing the analysis of their environment, neighboring genes.
- In order to subsequently identify the new alleles of existing genes, the best results associated with a given new strains’ genes are analyzed in terms of their L-V-GENE-UNIT (L-PART1 to V-RS), but also their recombination signals and the leader of the V-REGIONs.
- Following this analysis of the similarity between the new genes and IMGT annotated gene (the IMGT reference repertoire), blocks of similar genes are gradually built around the identified “anchors”.
- Genes located between identified blocks are treated as insertions.
Previous nomenclature page is still available as archive (19/09/2024) here.