Half-IG exchange
Human IgG4 form bispecific antibodies through a dynamic half-IG exchange process, that occurs naturally in vivo, in which half molecules recombine with half molecules (i.e. one heavy and one light chain) from other IgG4 [1].
This proccess is also called Fab-arm exchange but this name has been debated [2].
Half-IG exchange in human IgG4
Heavy chains (H) of human immunoglobulin IgG4 are covalently linked by two disulfide bridges located in their hinge region.
However, it has been shown that the two inter-heavy chain disulfide bonds of IgG4 are in equilibrium with the intra-chain disulfide bonds forms [3].
As a consequence, destabilizing the inter-heavy chain disulfide bonds between the heavy chains will favor the half-IG exchange.
In IgG4, two amino acids have been shown to play a key role in half-IG exchange: the serine (Ser) S10, located in the hinge region and the arginine (Arg) R88, located at the interface between the two CH3 domains.
S10, which is a Pro in IgG1, is thought to make the inter-heavy chains disulfide bridges more accessible to reducing conditions [4], while, R88, which is a lys in IgG1, has been shown to decrease the CH3-CH3 interaction, allowing human IgG4 to engage half-IG exchange [5].
The half-IG exchange event occurs also for therapeutic IgG4 antibodies.
Indeed, natalizumab has been shown to exchange half molecules with endogenous human IgG4 in natalizumab-treated patients.
Whereas, gemtuzumab carries a S10P mutation in its hinge region, making it less succeptible to half-IG exchange in vivo [6].

Figure 1. The 3D structure of the human IgG b12 antibody is shown with the hinge region (containing P10) and the CH3 domains (containing K88) labeled in red.
The heavy chains are represented in blue and purple, the light chains are in orange and green, respectively.
Strategy for the generation of bispecific IgG1 by controlled half-IG exchange
In this approach, the authors used matched amino acid change at the interface between the two CH3 domains (selected for their ability to decrease the interaction strength in both homodimers) in combination with a mild reducing agent, to favor efficient bispecific antibodies formation [7].
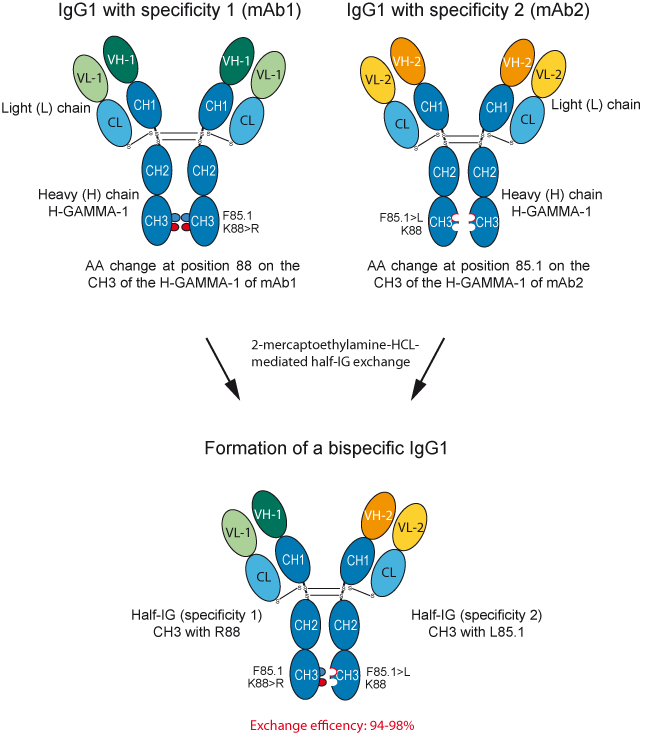
Figure 2. Strategy used in the controlled half-IG exchange.
'VH-1' and 'VL-1' represents the VH and VL domains of a first antibody with a given specificity, whereas 'VH-2' and 'VL-2' represents the VH and VL domains of a second antibody with a different specificity.
The two antibodies, each containing single matched amino acid change in CH3 are expressed separately and recombined using the reducing agent to favor half-IG exchange. The efficency of half-IG exchange ranges between 94 and 98% [7].
Molecular interactions at the interface between the two CH3 domains
In the interface between the CH3 domains, the lysine (Lys) K88 in strand E is within close proximity with the phenylalanine (Phe) F85.1 in strand E on the partner CH3 domain.
K88 makes numerous van der Waals contacts with F85.1 (red box). However, K88 also makes numerous contacts with L24, K26, D84.2 and Y86 on the other domain (black arrows).
Table 1. Interactions involving K88 of the CH3 domain of IgG1 H chains.
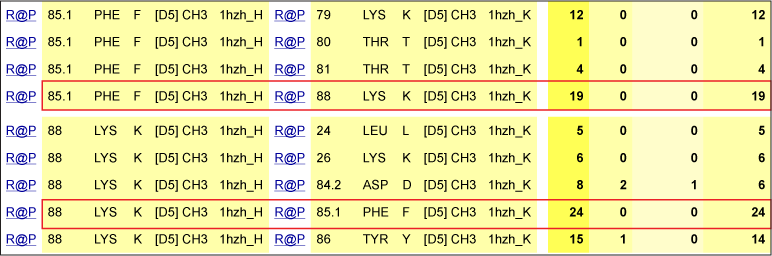
IMGT Contact analysis are from the human IGHG1 (1hzh in IMGT/3Dstructure-DB).
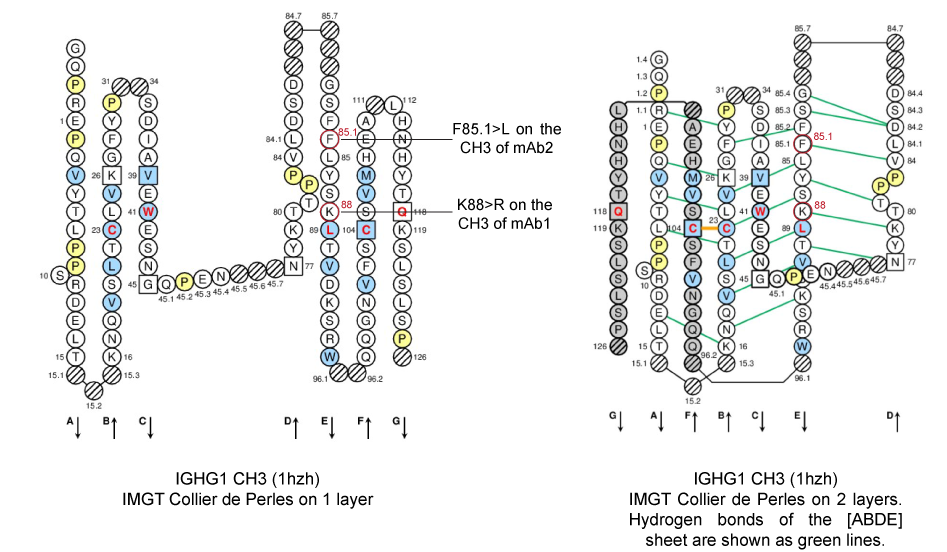
Figure 3. IMGT Collier de Perles from human IGHG1 (1hzh, IMGT/3Dstructure-DB).
Positions K88 and F85.1 of the CH3-DOMAIN are shown in the IMGT Collier de Perles, in red circles. K88 and F85.1 (strand E) are in the [ABDE] sheet at the interface between the 2 CH3 domains.
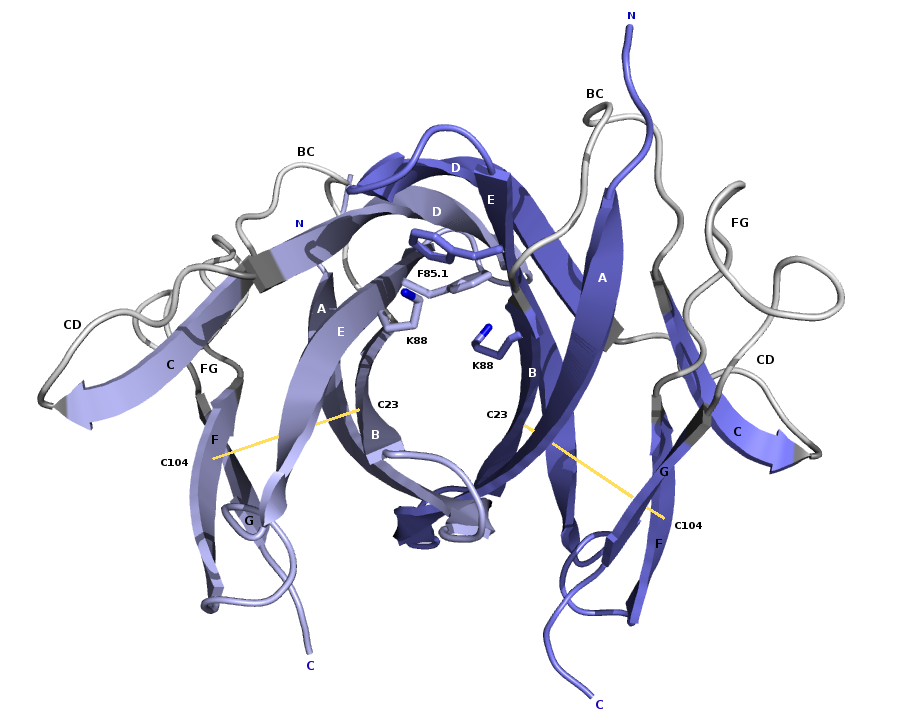
Figure 4. 3D structure of the CH3 dimer interface from the human IGHG1 (1hzh in IMGT/3Dstructure-DB).
The residues K88 and F85.1 involved in the half-IG exchange are represented.
In their work, the matched pair was created by the amino acid changes of K88>R in the first CH3 domain and F85.1>L on the partner CH3 domain [6].
| [1] | van der Neut Kolfschoten, M. et al. Sciences, 317(5844):1554-7 (2007) PMID: 17872445 |
| [2] | Schuurman, J. et al. mAbs 4(6):636 (2012) PMID: 22955209 |
| [3] | Schuurman, J. et al. Mol Immunol. 38(1):1-8.(2001) PMID: 11483205 |
| [3] | Bloom, J. W. et al. Protein Sci. 6(2):407-15 (1997) PMID: 9041643 |
| [4] | Labrijn, A. F. et al. J Immunol. 187(6):3238-46 (2011) PMID: 21841137 |
| [5] | Labrijn, A. F. et al. Nat Biotechnol. 27(8):767-71 (2009) PMID: 19620983 |
| [6] | Labrijn, A. F. et al. Proc Natl Acad Sci U S A. 110(13):5145-50 (2013) PMID: 23479652 |
Last updated:
Authors: Souphatta Sasorith and Marie-Paule Lefranc
Editor: Chantal Ginestoux
IMGT Home page |
IMGT Repertoire (IG and TR) |
IMGT Repertoire (MH) |
IMGT Repertoire (RPI) |
IMGT Index |
IMGT Scientific chart |
IMGT Education |
IMGT Latest news ![]()



© Copyright 1995-2026 IMGT®, the international ImMunoGeneTics information system® | Terms of use | About us | Contact us | Citing IMGT

