IMGT/3Dstructure-DB contact analysis
- Atom contact types
- Hydrogen bond angle assessment
- Atom contact categories
- Domain/chain level contacts
The contact analysis in IMGT/3Dstructure-DB [1] is performed with a home made program. Two atoms are in contact when no water molecule can take place between them.
Atom contact types
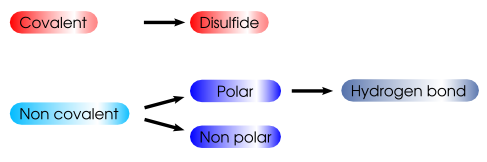
A contact is considered to be a covalent link if the distance between their centers is less than the sum of the Van Der Waals radius of the two atoms. If the two atoms are sulfurs, the contact is a disulfide bridge. A non covalent contact is either a polar contact, if the two atoms are azote and/or nitrogen, or a non polar contact. A polar contact is defined as an hydrogen bond if the distance between the atom centers is less than 3.5 Å and the hydrogen bond angles are correct (as described below). An hydrogen bond contact counts also for a polar contact and for a non covalent contact.
 The total number of atom contacts is the sum of the number of covalent
links and non covalent contacts. The number of non covalent contacts is the sum of the number of polar and non polar contacts.
The number of hydrogen bonds is less than the number of polar contacts.
The total number of atom contacts is the sum of the number of covalent
links and non covalent contacts. The number of non covalent contacts is the sum of the number of polar and non polar contacts.
The number of hydrogen bonds is less than the number of polar contacts.
Hydrogen bond angle assessment
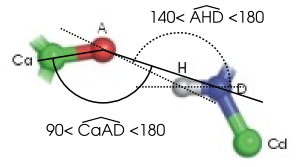
| Ca | Carbon link to the acceptor atom | H | Hydrogen | |
|---|---|---|---|---|
| A | Acceptor atom | D | Donor atom | |
| Cd | Carbon link to the donor atom |
The angle between A, H and D must be greater than 140° (and less than 180°) and the angle between Ca, A and D must be greater than 90° (and less than 180°).
For nitrogen hydrogen donors that are linked to two heavy atoms, the hydrogen position is explicitly calculated because its position is directly derived from the known position of heavy atoms.
For other hydrogen donors, the hydrogen position is not directly calculated. Cd, D, H and A are supposed to be coplanar, that is an ideal figure for having an hydrogen bond which is always possible considering the free dihedral rotation of the Cd-D link. The distance DH and the angle CdDH are taken from a library (AMBER) and triangle angle relations allows the calculation of the AHD and CaAD angles.
Atom contact categories
Atom contact categories are:
- backbone/backbone contacts (BB)
- sidechain/sidechain contacts (SS)
- backbone/sidechain contacts (BS)
- sidechain/backbone contacts (SB)
In IMGT/3Dstructure-DB [1] atom contacts per category are counted at the residue level.
Domain/chain level contacts
Atom contact types and categories are used IMGT structural analysis [2-4] to analyse each domain/chain and domain/domain interface. This analysis provides:
- the number of residues implicated (total and number per partner)
- the number of atom contacts between residues
- the sum of the atom contacts per type and per category
| [1] | Kaas, Q., Lefranc, M.-P. IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acid Res., 32:D208-D210 (2004). PMID: 14681396, LIGM:281. |
| [2] | Kaas, Q., Lefranc, M.-P. 'IMGT/StructuralQuery: a tool for structural data analysis of immunoglobulin and T cell receptor variable domains' ECCB 2003 |
| [3] | Kaas, Q., Lefranc, M.-P. 'IMGT/3Dstructure-DB for immunoglobulin, T cell receptor and MHC structural data' ECCB 2002 |
| [4] | Kaas, Q. and Lefranc M.-P. 'T cell receptor/peptide/MHC molecular characterization and standardized pMHC contact sites in IMGT/3Dstructure-DB' In Silico Biology 5:0046 (2005) |



