Here you are: IMGT Web resources > IMGT Education
Polyadenylation (or Poly(A)) signal, site and tail
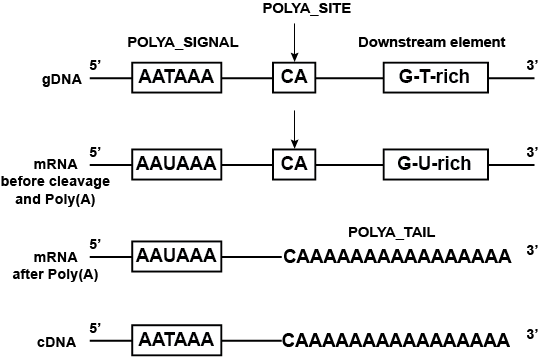
The polyadenylation or Poly(A) is the process required for the synthesis of messenger RNA (mRNA) in which an endonucleolityc RNA cleavage is coupled with synthesis of polyadenosine monophosphate (adenine base) on the newly formed 3' end. The sequence elements for polyadenylation include the polyadenylation signal (POLYA_SIGNAL) and the polyadenylation site (POLYA_SITE). In mRNA or cDNA the added stretch of polyadenosine monophosphate is the polyadenylation tail (POLYA_TAIL).
Polyadenylation signal
- Poly(A) signal
- IMGT label: POLYA_SIGNAL
- Conserved motif: AATAAA
- The polyadenylation signals are located downstream of the 3' exons [1].
- Example: A POLYA_SIGNAL located 103 bp downstream of the human IGHG3 CH3-CHS
exon is used in the transcription of
secreted gamma3 chains.
A POLYA_SIGNAL located downstream of the human IGHG3 M2 exon is used in the transcription of membrane gamma3 chains [2].
Polyadenylation site
- Poly(A) site
- IMGT label: POLYA_SITE
- The polyadenylation site is the site of cleavage at which POLYA_TAIL is added in mRNA. It is localized downstream of the POLYA_SIGNAL. The POLYA_SITE can be determined by comparing cDNA and gDNA.
- The sequence at/or immediately 5' to the site of RNA cleavage is frequently (but not always) CA. A 'G-U-rich' element or Downstream element usually lies just downstream of the POLYA_SITE in gDNA, which is important for efficient processing. In those cases, the POLYA_SITE maybe located in gDNA.[3]
Polyadenylation tail
- Poly(A) tail
- IMGT label: POLYA_TAIL
- Stretch of adenosine monophosphate (with only adenine bases) at the 3' end of mRNA or cDNA.
| [1] | Wahle E. and Keller W., Ann. Rev. Biochem., 61:419-440 (1992) |
| [2] | Lefranc, M.-P. and Lefranc, G., The immunoglobulin FactsBook, Academic Press, London, 458 pages (2001) ISBN: 012441351X |
| [3] | Manley J.-L. and Takagaki Y., Science, 274:1481-1482 (1996) |



