WELCOME!
to IMGT/Collier-de-Perles Documentation
IMGT®, the international ImMunoGeneTics information system®

WELCOME!
to IMGT/Collier-de-Perles Documentation
IMGT®, the international ImMunoGeneTics information system®

Ruiz, M. and Lefranc, M.-P. Immunogenetics, 53:857-883 (2002). PMID:11862387
Kaas, Q. and Lefranc, M.-P.
Current Bioinformatics, 2:21-30 (2007).
![]()
Kaas, Q., Ehrenmann, F. and Lefranc, M.-P.
Brief. Funct. Genomic Proteomic, 6:253-264 (2007).
PMID:18208865
![]()
Ehrenmann, F., Giudicelli., V, Duroux, P., Lefranc, M.-P.
Cold Spring Harbor Protoc., 6:726-736 (2011).
PMID:21632776
Abstract
also in IMGT booklet with generous provision from Cold Spring Harbor (CSH) Protocols
![]() (high res)
(high res)
![]() (low res)
(low res)
IMGT/Collier-de-Perles is part of IMGT®, the international ImMunoGeneTics information system®, the high-quality integrated information system specialized in immunoglobulins (IG), T cell receptors (TR), major histocompatibility complex (MH) of human and other vertebrates species, immunoglobulin superfamily (IgSF), MHC superfamily (MhcSF) and related proteins of the immune system (RPI), created in 1989 by Marie-Paule Lefranc (Laboratoire d'ImmunoGénétique Moléculaire, LIGM, Université Montpellier II and CNRS) and on the Web since July 1995.
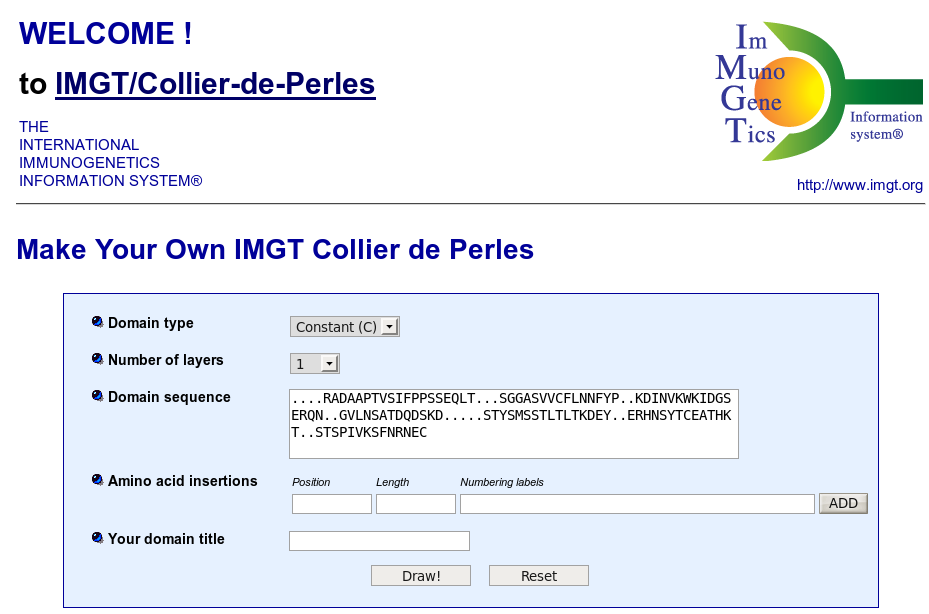
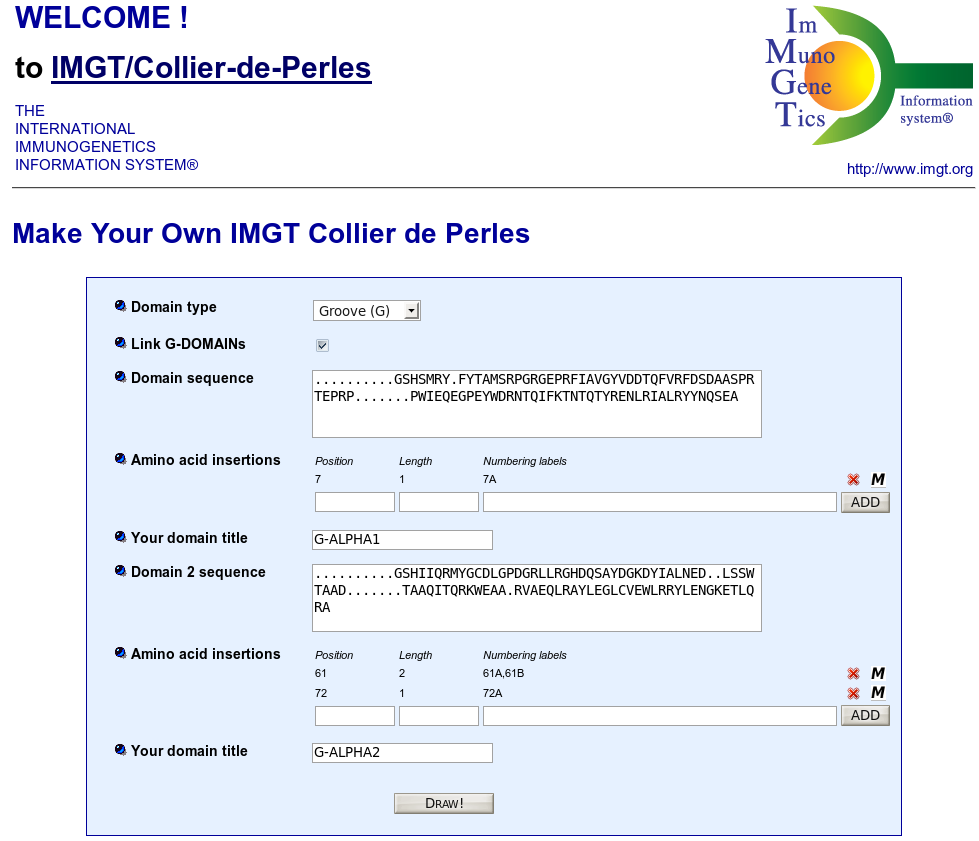
IMGT/Collier-de-Perles is a tool that allows to draw standardized IMGT 2D graphical representations of protein domains, or IMGT Colliers de Perles, starting from the user own domain amino acid sequences.
IMGT Colliers de Perles can be drawn for four domain types:
By default, the web application is loaded for V type domains.
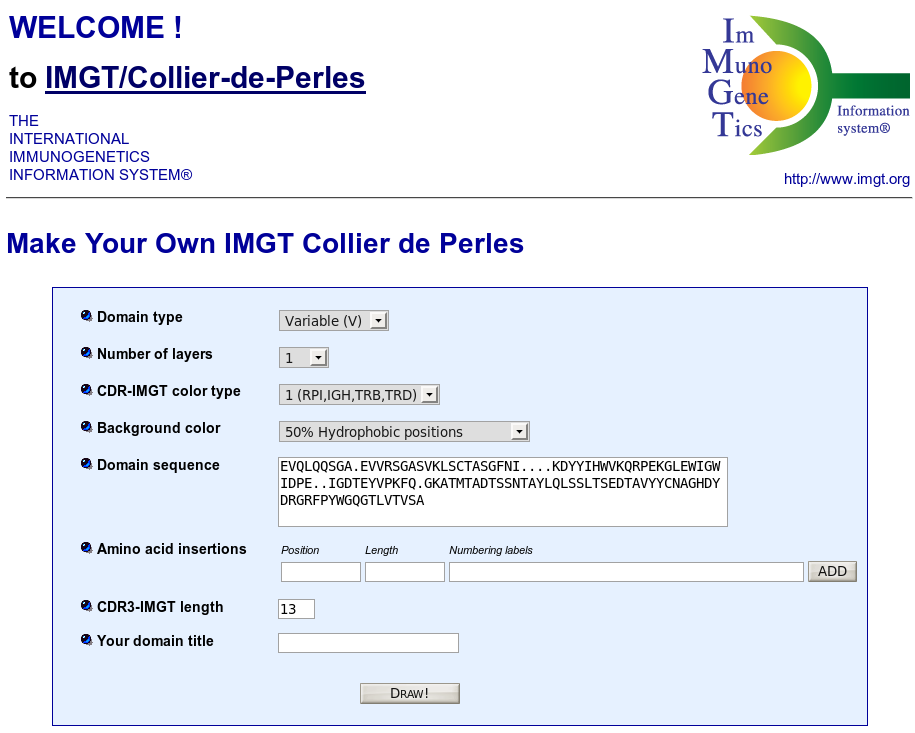
EVQLQQSGA.EVVRSGASVKLSCTASGFNI....KDYYIHWVKQRPEKGLEWIGWIDPE..IGDTEYVPKFQ.GKATMTADTSSNTAYLQLSSLTSEDTAVYYCNAGHDYDRGRFPYWGQGTLVTVSAOr
.KEVEQNSGPLSVPEGAIASLNCTYSDRG......SQSFFWYRQYSGKSPELIMSIYS....NGDKED.....GRFTAQLNKASQYVSLLIRDSQPSDSATYLCAVTTDS..WGKLQFGAGTQVVVTP
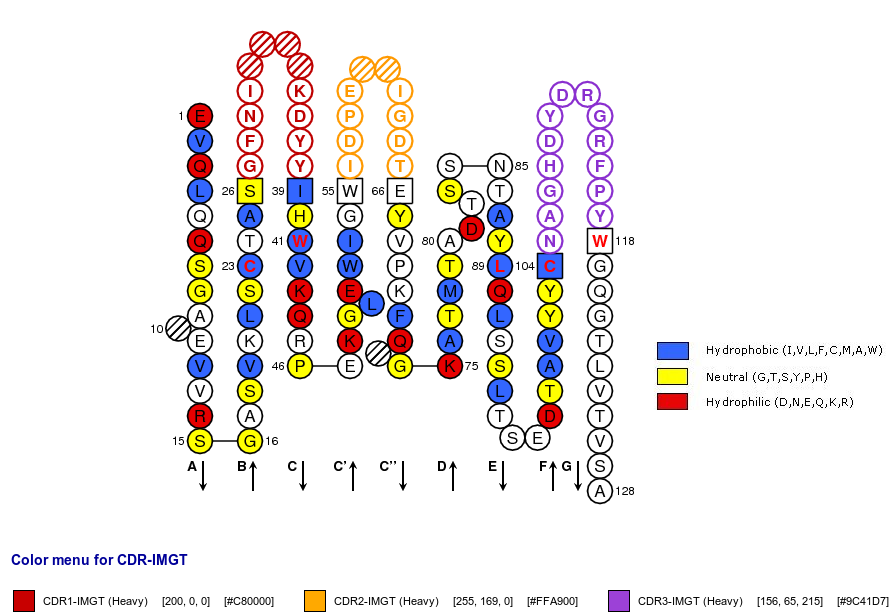
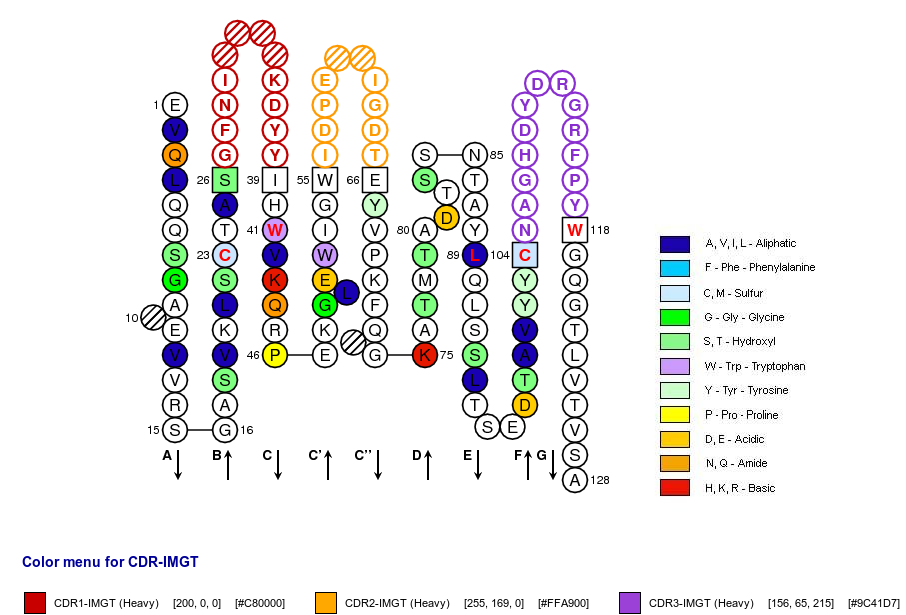
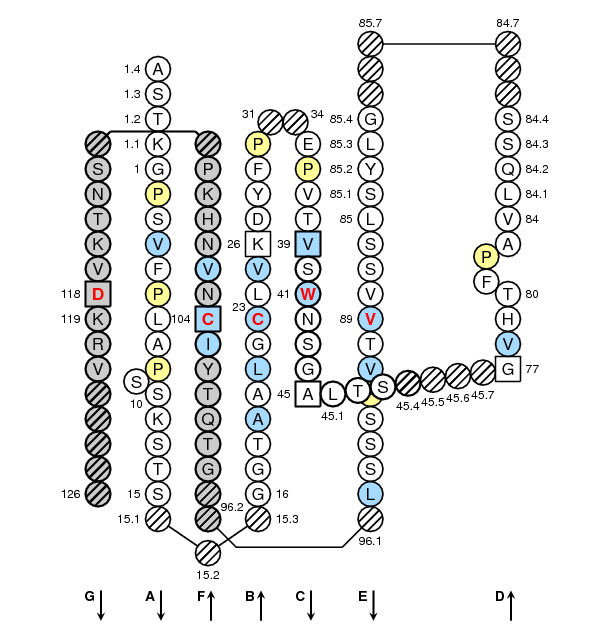
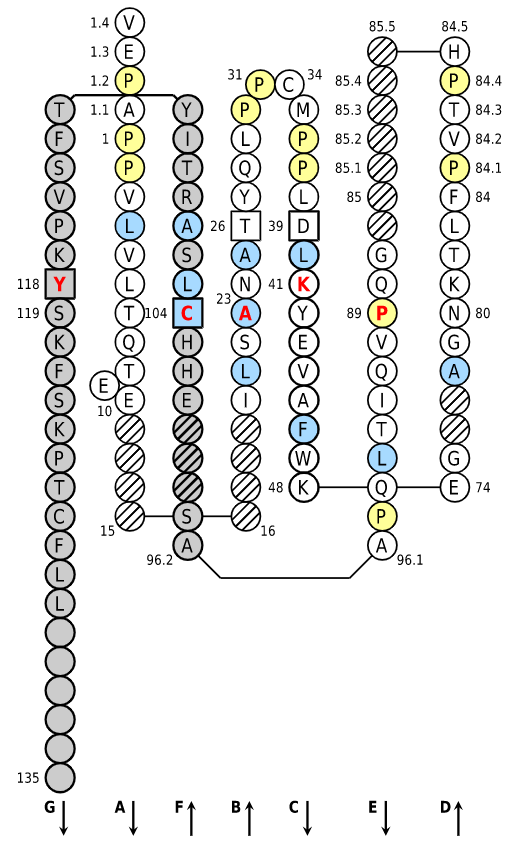
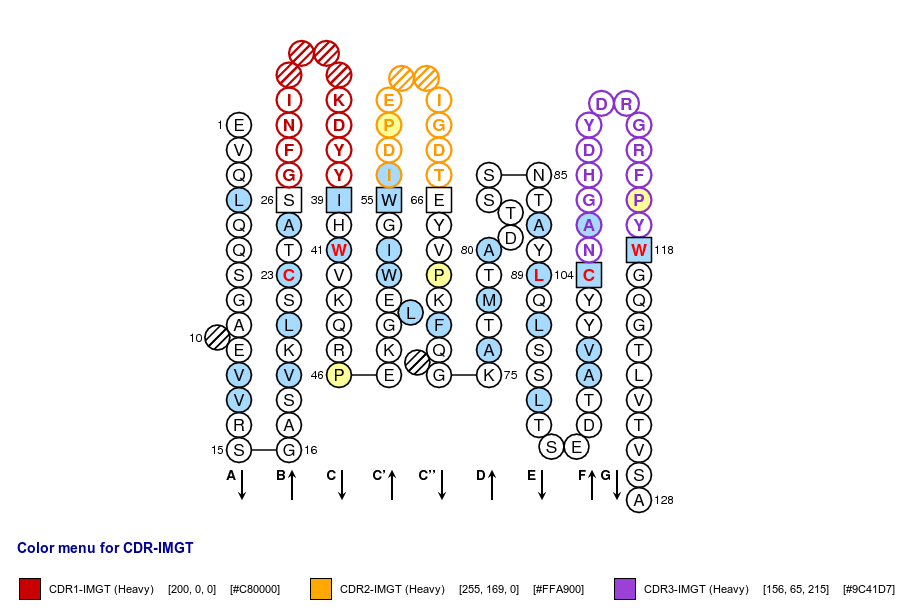 Positions in blue mean that the AA of the user sequence at these positions is hydrophobic (hydropathy index with positive value) or is a tryptophan (W), like in 50% or more of analysed V domains. Positions with red and bold letters indicate the five conserved positions of a V domain: 1st-CYS 23, CONSERVED-TRP 41, hydrophobic 89, 2nd-CYS 104 and J-TRP 118. Anchor positions are in square. Hatched positions correspond to gaps according to the IMGT unique numbering for V domain. Proline are shown in yellow. Arrows indicate the beta strands and their direction. The CDR-IMGT lengths of this domain are [10.7.13].
Positions in blue mean that the AA of the user sequence at these positions is hydrophobic (hydropathy index with positive value) or is a tryptophan (W), like in 50% or more of analysed V domains. Positions with red and bold letters indicate the five conserved positions of a V domain: 1st-CYS 23, CONSERVED-TRP 41, hydrophobic 89, 2nd-CYS 104 and J-TRP 118. Anchor positions are in square. Hatched positions correspond to gaps according to the IMGT unique numbering for V domain. Proline are shown in yellow. Arrows indicate the beta strands and their direction. The CDR-IMGT lengths of this domain are [10.7.13].
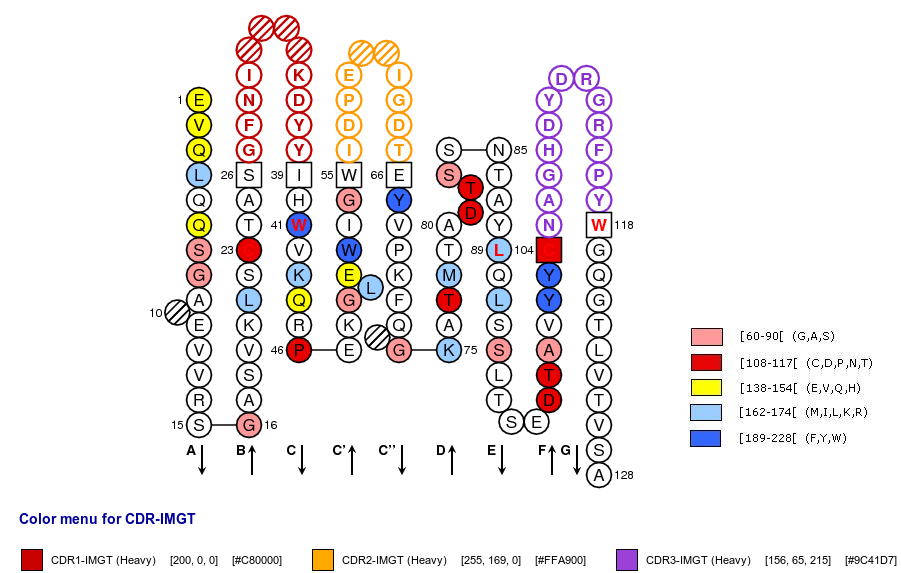
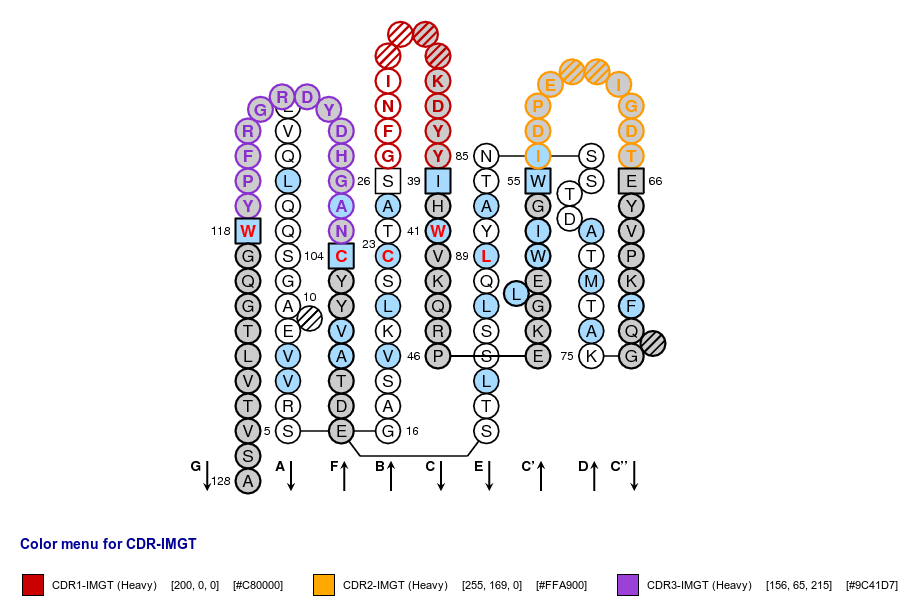
 IMGT Collier de Perles of a variable (V) domain with "IGH 80% hydropathy positions". Colored positions (3 classes) mean that the property of the AA of the user sequence at these positions is found in more than 80% of the analysed IGH V domains.
IMGT Collier de Perles of a variable (V) domain with "IGH 80% hydropathy positions". Colored positions (3 classes) mean that the property of the AA of the user sequence at these positions is found in more than 80% of the analysed IGH V domains.
GDQVEQSPSALSLHEGTDSALRCNFTTTM.......RSVQWFRQNSRGSLISLFYLAS.....GTKEN.....GRLKSAFDSKERRYSTLHIRDAQLEDSGTYFCAADTWHISEGYELGTDKLVFGQGTQVTVEP
....ASTKGPSVFPLAPSSKSTS...GGTAALGCLVKDYFP..QPVTVSWNSGALTS....GVHTFPAVLQSS......GLYSLSSVVTVPSSSL...GTQTYICNVNHKP..SNTKVDKKVOr
....RADAAPTVSIFPPSSEQLT...SGGASVVCFLNNFYP..KDINVKWKIDGSERQN..GVLNSATDQDSKD.....STYSMSSTLTLTKDEY..ERHNSYTCEATHKT..STSPIVKSFNRNEC
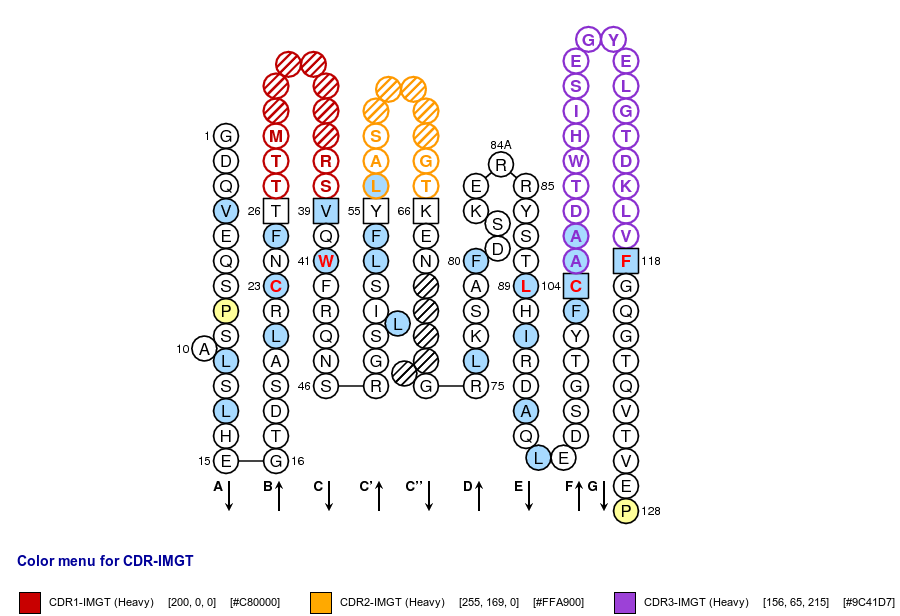
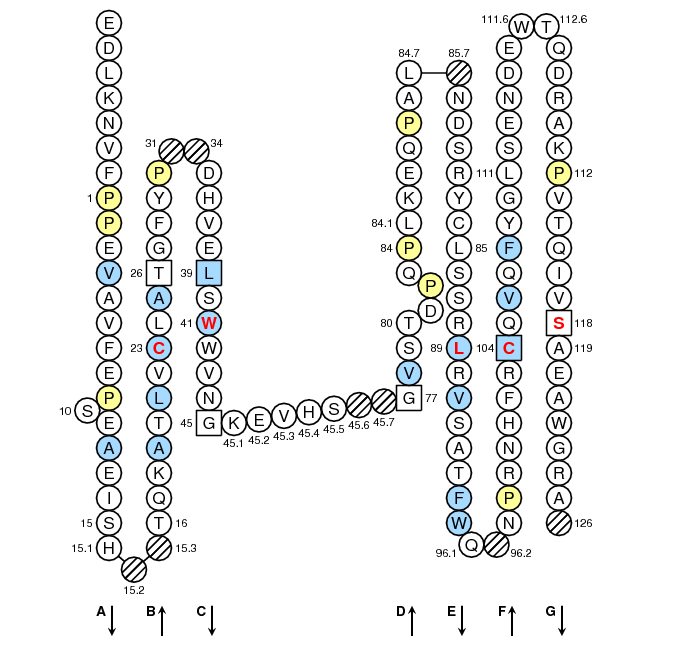
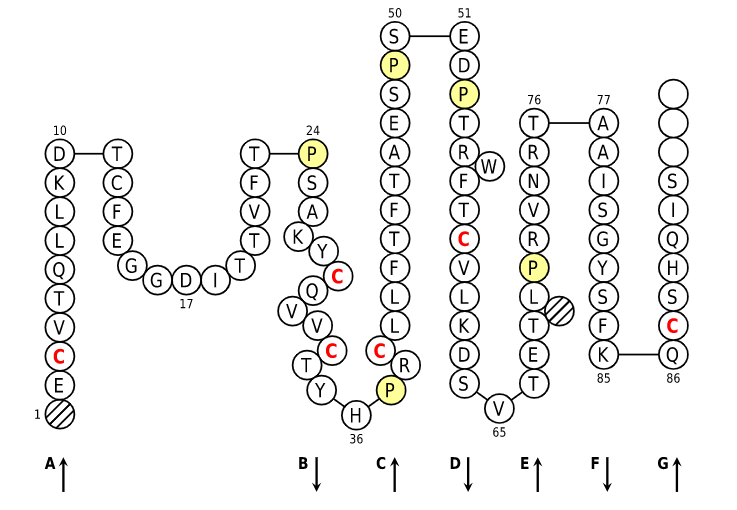
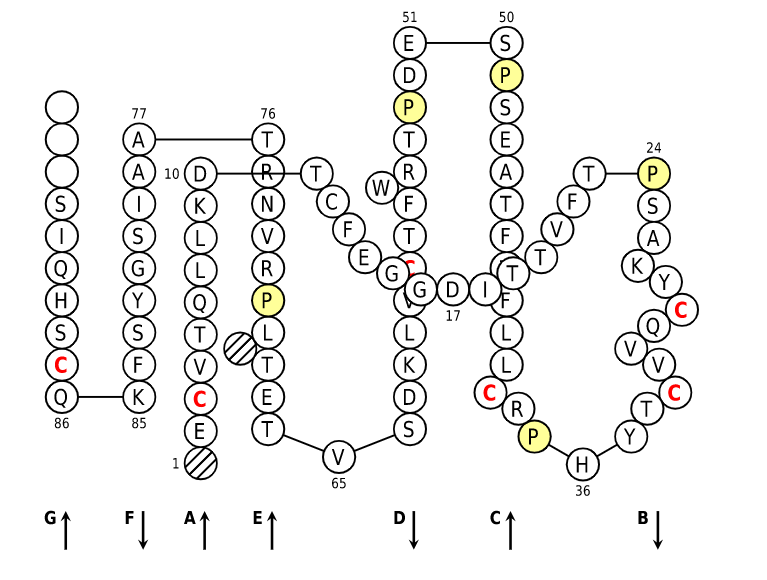
 Positions in blue mean that the AA of the user sequence at these positions is hydrophobic (hydropathy index with positive value) or is a tryptophan (W), like in 50% or more of analysed V domains. Positions with red and bold letters indicate the five conserved positions of a V domain: 1st-CYS 23, CONSERVED-TRP 41, hydrophobic 89 and 2nd-CYS 104. Anchor positions are in square. Hatched positions correspond to gaps according to the IMGT unique numbering for C domain. Proline are shown in yellow. Arrows indicate the beta strands and their direction.
Positions in blue mean that the AA of the user sequence at these positions is hydrophobic (hydropathy index with positive value) or is a tryptophan (W), like in 50% or more of analysed V domains. Positions with red and bold letters indicate the five conserved positions of a V domain: 1st-CYS 23, CONSERVED-TRP 41, hydrophobic 89 and 2nd-CYS 104. Anchor positions are in square. Hatched positions correspond to gaps according to the IMGT unique numbering for C domain. Proline are shown in yellow. Arrows indicate the beta strands and their direction.
.EDLKNVFPPEVAVFEPSEAEISH..TQKATLVCLATGFYP..DHVELSWWVNGKEVHS..GVSTDPQPLKEQPAL.NDSRYCLSSRLRVSATFWQ.NPRNHFRCQVQFYGLSENDEWTQDRAKPVTQIVSAEAWGRA
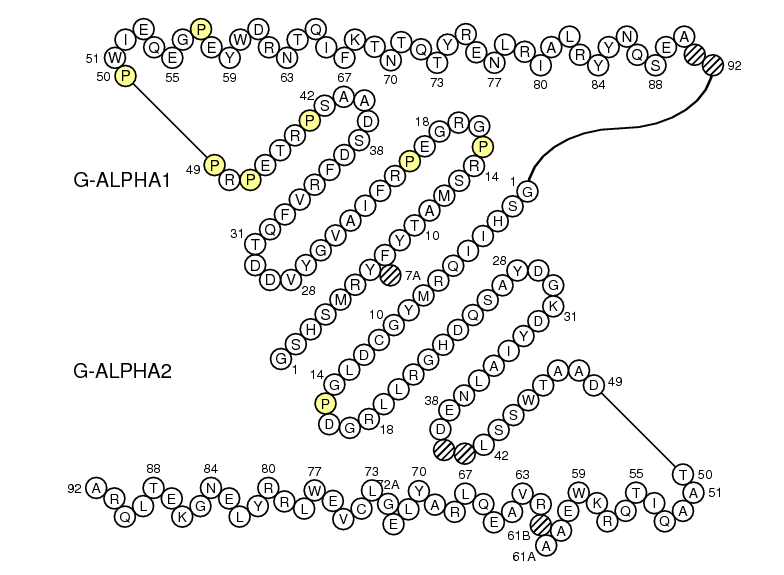
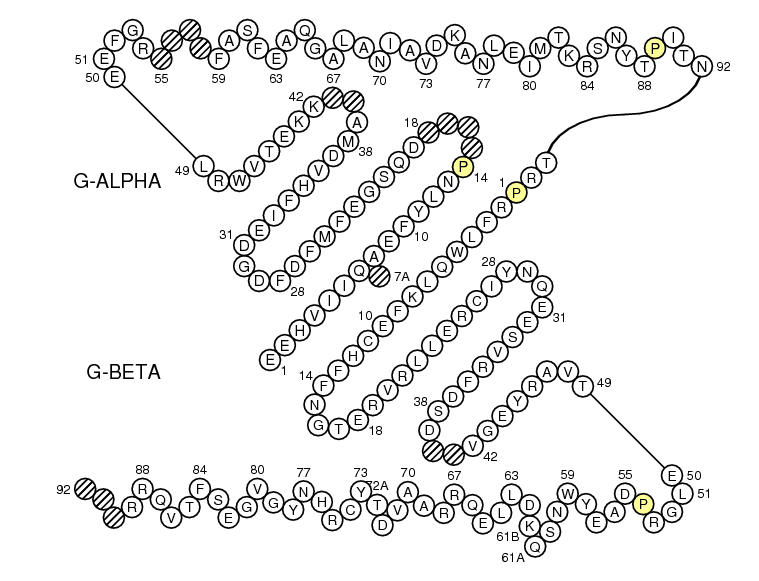
G-ALPHA1 GSHSMRY.FSAAVSRPGRGEPRFIAMGYVDDTQFVRFDSDSACPRMEPRA.......PWVEQEGPEYWE..EETRNTKAHAQ.TDRMNLQTLRGYYNQSEAOr
G-ALPHA ..........EEHVIIQ.AEFYLNP....DQSGEFMFDFDGDEIFHVDMA..KKETVWRL.......EEFGR....FASFEAQGALANIAVDKANLEIMTKRSNYTPITN
G-ALPHA2 ..........GSHIIQRMYGCDLGPDGRLLRGHDQSAYDGKDYIALNED..LSSWTAAD.......TAAQITQRKWEAA.RVAEQLRAYLEGLCVEWLRRYLENGKETLQRAOr
G-BETA ........TRPRFLWQLKFECHFFNGTERVRLLERCIYNQEESVRFDSD..VGEYRAVT.......ELGRPDAEYWNSQKDLLEQRRAAVDTYCRHNYGVGESFTVQRR
.........GFQPKVQSRLVGGSSICEGTVEVRQGAQWAALCDSSSARSSLRWEEVCREQQCGSVNSYRVLDAGDP..........TSRGLFCPHQK......LSQCHELWER....NSYCKKVFVTCQ..
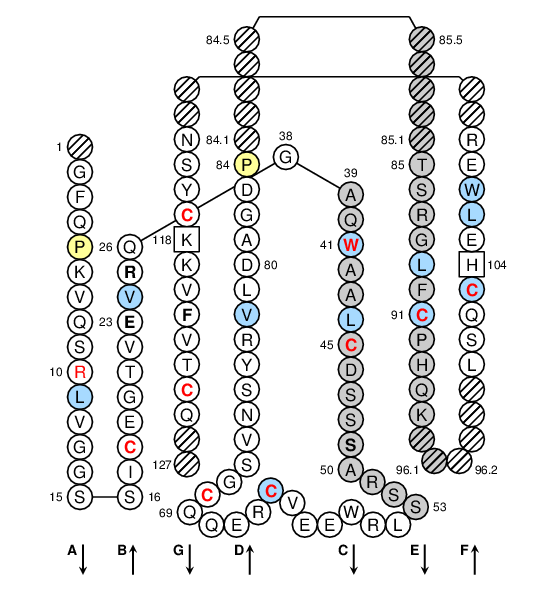
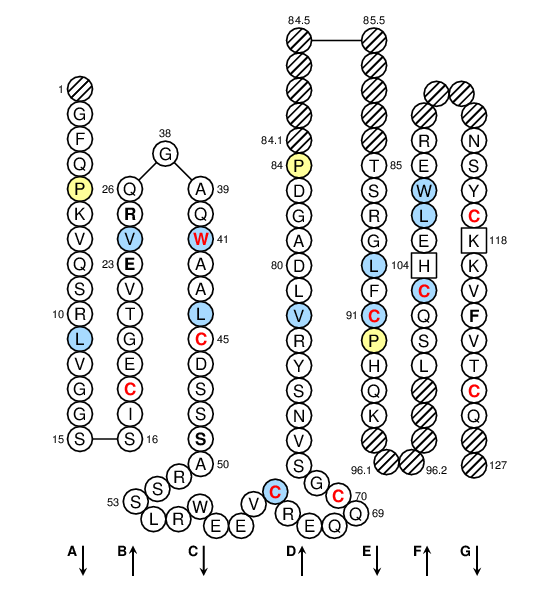 Positions in blue mean that the AA of the user sequence at these positions is hydrophobic (hydropathy index with positive value) or is a tryptophan (W), like in 50% or more of analysed S domains. Positions with red and bold letters indicate the five conserved positions of a S domain: CYS 18, CONSERVED-TRP 41, CYS 45, CYS 60, CYS 70, hydrophobic 89, CYS 91, CYS 103, CYS 117 and CYS 124. Hatched positions correspond to gaps according to the IMGT unique numbering for S domain. Proline are shown in yellow. Arrows indicate the beta strands and their direction.
Positions in blue mean that the AA of the user sequence at these positions is hydrophobic (hydropathy index with positive value) or is a tryptophan (W), like in 50% or more of analysed S domains. Positions with red and bold letters indicate the five conserved positions of a S domain: CYS 18, CONSERVED-TRP 41, CYS 45, CYS 60, CYS 70, hydrophobic 89, CYS 91, CYS 103, CYS 117 and CYS 124. Hatched positions correspond to gaps according to the IMGT unique numbering for S domain. Proline are shown in yellow. Arrows indicate the beta strands and their direction.

Created: 10/12/06
Editors: Manuel Ruiz (2001), Quentin Kaas (2001-2005), François Ehrenmann (2006-2010), and Patrice Duroux (since 2011)