Frequently asked questions
IMGT/GENE-DB
- Is it possible to retrieve flanking sequence at the 5' and/or 3' ends of IMGT labels that describe IMGT/GENE-DB annotated sequences?
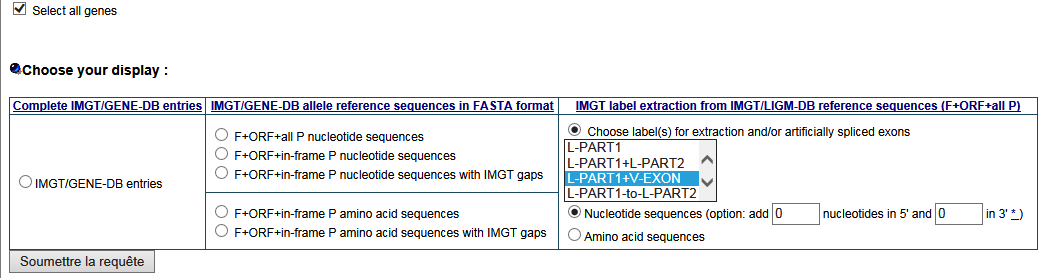
- How can I retrieve the V leader sequences from IMGT reference sequences?
- How to obtain from IMGT/GENE-DB complete amino acid sequences (artificially spliced) of a constant gene, or of a group of C genes?
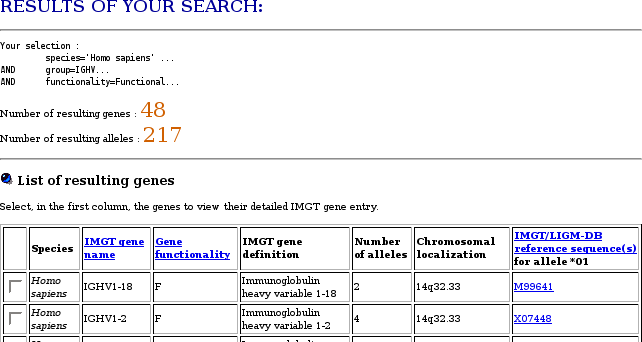
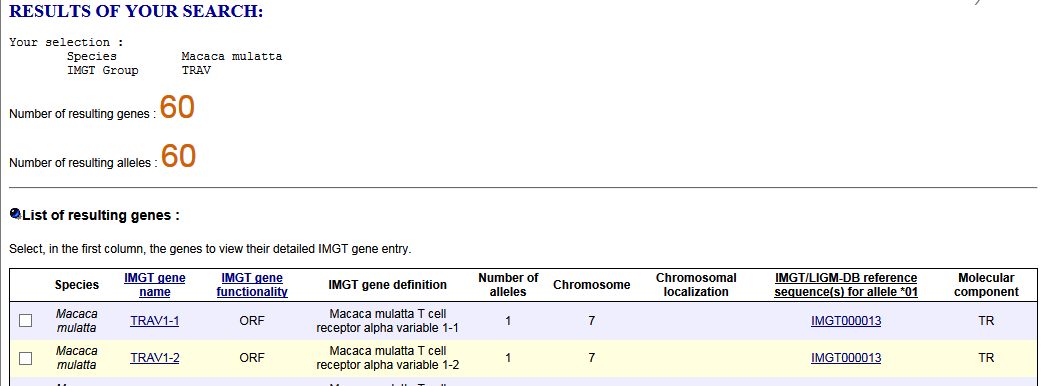
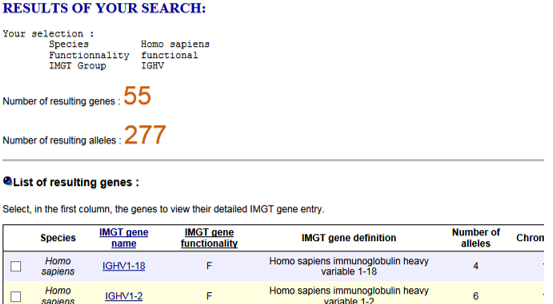
- How to download the sequences of the available IG or TR genes of a given group of a given species, for example Homo sapiens IGHV, from IMGT/GENE-DB?
- How to retrieve sequences of the alleles of the human constant genes?
- How to get nucleotide sequences of the human constant IG genes for primer design?
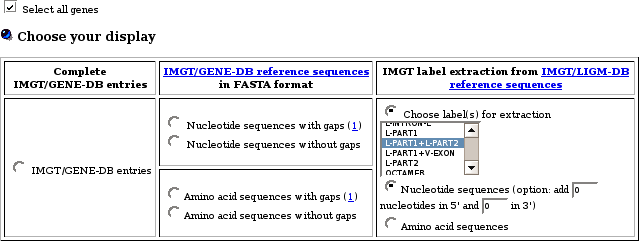
- How can I export FASTA sequences for V genes that include the L-REGION in addition to V-REGION?
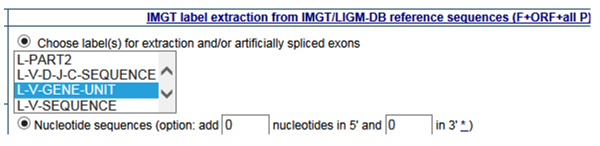
- How can I export FASTA sequences for L-V GENE-UNIT?