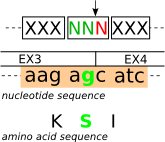Color menu for amino acids
Color menu for amino acids is according to [1]
| |
[230,6,6] |
[#E60606] |
R - Arg - Arginine |
| |
[198,66,0] |
[#C64200] |
K - Lys - Lysine |
| |
[255,102,0] |
[#FF6600] |
Q - Gln - Glutamine |
| |
[255,153,0] |
[#FF9900] |
N - Asn - Asparagine |
| |
[255,204,0] |
[#FFCC00] |
E - Glu - Glutamic Acid |
| |
[255,204,153] |
[#FFCC99] |
D - Asp - Aspartic Acid |
| |
[255,255,153] |
[#FFFF99] |
H - His - Histidine |
| |
[255,255,0] |
[#FFFF00] |
P - Pro - Proline |
| |
[204,255,204] |
[#CCFFCC] |
Y - Tyr - Tyrosine |
| |
[204,153,255] |
[#CC99FF] |
W - Trp - Tryptophan |
| |
[204,255,153] |
[#CCFF99] |
S - Ser - Serine |
| |
[0,255,153] |
[#00FF99] |
T - Thr - Threonine |
| |
[0,255,0] |
[#00FF00] |
G - Gly - Glycine |
| |
[204,255,255] |
[#CCFFFF] |
A - Ala - Alanine |
| |
[153,204,255] |
[#99CCFF] |
M - Met - Methionine |
| |
[0,255,255] |
[#00FFFF] |
C - Cys - Cysteine |
| |
[0,204,255] |
[#00CCFF] |
F - Phe - Phenylalanine |
| |
[51,102,255] |
[#3366FF] |
L - Leu - Leucine |
| |
[0,0,255] |
[#0000FF] |
V - Val - Valine |
| |
[0,0,128] |
[#000080] |
I - Ile - Isoleucine |
Color menu for the 11 IMGT amino acid Physicochemical classes
Color menu for the 11 IMGT amino acid Physicochemical classes is according to [2]
| |
[27,4,172] |
[#1B04AC] |
A,I,L,V- Aliphatic |
| |
[255,255,0] |
[#FFFF00] |
P - Pro - Proline |
| |
[204,255,204] |
[#CCFFCC] |
Y - Tyr - Tyrosine |
| |
[204,153,255] |
[#CC99FF] |
W - Trp - Tryptophan |
| |
[0,255,0] |
[#00FF00] |
G - Gly - Glycine |
| |
[0,204,255] |
[#00CCFF] |
F - Phe - Phenylalanine |
| |
[204,236,255] |
[#CCECFF] |
C,M - Sulfur |
| |
[137,248,139] |
[#89F88B] |
S,T - Hydroxyl |
| |
[255,204,0] |
[#FFCC00] |
D,E - Acidic |
| |
[204,165,4] |
[#CCA504] |
N,Q - Amide |
| |
[236,21,4] |
[#EC1504] |
R,H,K - Basic |
Color menu for the 7 IMGT amino acid Chemical classes
If IMGT tools are limited to 7 choices of colors, the following menu is used.
| |
[27,4,172] |
[#1B04AC] |
A,G,I,L,P,V- Aliphatic |
| |
[204,236,255] |
[#CCECFF] |
C,M - Sulfur |
| |
[137,248,139] |
[#89F88B] |
S,T - Hydroxyl |
| |
[255,204,0] |
[#FFCC00] |
D,E - Acidic |
| |
[204,165,4] |
[#CCA504] |
N,Q - Amide |
| |
[236,21,4] |
[#EC1504] |
R,H,K - Basic |
| |
[204,153,255] |
[#CC99FF] |
F,W,Y - Aromatic |
Color menu for the 5 IMGT amino acid Volume classes
Color menu for the 5 IMGT amino acid Volume classes is according to [2]
| |
[255,153,153] |
[#FF9999] |
[60-90[ (A,G,S) |
| |
[230,6,6] |
[#E60606] |
[108-117[ (N,D,C,P,T) |
| |
[255,255,0] |
[#FFFF00] |
[138-154[ (Q,E,H,V) |
| |
[153,204,255] |
[#99CCFF] |
[162-174[ (R,I,L,K,M) |
| |
[51,102,255] |
[#3366FF] |
[189-228[ (F,W,Y) |
Color menu for the 3 IMGT amino acid Hydropathy classes
Color menu for the 3 IMGT amino acid Hydropathy classes is according to [2]
| |
[51,102,255] |
[#3366FF] |
A,C,I,L,M,F,W,V - Hydrophobic |
| |
[255,255,0] |
[#FFFF00] |
G,H,P,S,T,Y - Neutral |
| |
[230,6,6] |
[#E60606] |
R,N,D,Q,E,K - Hydrophilic |
Color menu for the 3 IMGT amino acid Charge classes
Color menu for the 3 IMGT amino acid Charge classes is according to [2]
| |
[204,236,255] |
[#CCECFF] |
A,N,C,Q,G,I,L,M,F,P,S,T,W,Y,V - Uncharged |
| |
[236,21,4] |
[#EC1504] |
R,H,K - Positive charged |
| |
[255,204,0] |
[#FFCC00] |
D,E - Negative charged |
Color menu for the 2 IMGT amino acid Polarity classes
Color menu for the 2 IMGT amino acid Polarity classes is according to [2]
| |
[230,6,6] |
[#E60606] |
R, N, D, Q, E, H, K, S, T, Y - Polar |
| |
[51,102,255] |
[#3366FF] |
A, C, G, I, L, M, F, P, W, V - Nonpolar |
Color menu for the 4 IMGT amino acid Hydrogen donor or acceptor atoms classes
Color menu for the 4 IMGT amino acid Hydrogen donor or acceptor atoms classes is according to [2]
| |
[0,204,255] |
[#00CCFF] |
R,K,W - Donor |
| |
[255,255,0] |
[#FFFF00] |
D,E - Acceptor |
| |
[137,248,139] |
[#89F88B] |
N,Q,H,S,T,Y - Donor and acceptor |
| |
[211,211,211] |
[#D3D3D3] |
A,C,G,I,L,M,F,P,V - None |
Color menu for nucleotides
| |
[0,204,0] |
[#00CC00] |
A |
| |
[204,0,0] |
[#CC0000] |
T |
| |
[255,179,0] |
[#FFB300] |
G |
| |
[0,0,204] |
[#0000CC] |
C |
Color menu for CDR-IMGT
| IG heavy, TR beta, TR delta chains |
| |
[200,0,0] |
[#C80000] |
CDR1-IMGT |
| |
[255,169,0] |
[#FFA900] |
CDR2-IMGT |
| |
[156,65,215] |
[#9C41D7] |
CDR3-IMGT |
| IG light, TR alpha, TR gamma chains |
| |
[96,96,228] |
[#6060E4] |
CDR1-IMGT |
| |
[70,213,0] |
[#46D500] |
CDR2-IMGT |
| |
[63,157,63] |
[#3F9D3F] |
CDR3-IMGT |
Color menu for genes
The color menu for genes is used in Locus representations.
| |
[0,238,0] |
[#00EE00] |
V-GENE Functional |
| |
[255,255,0] |
[#FFFF00] |
V-GENE ORF |
| |
[255,51,0] |
[#FF3300] |
V-GENE Pseudogene |
| |
[169,169,169] |
[#A9A9A9] |
V-GENE Vestigial |
| |
[0,0,228] |
[#0000E4] |
D-GENE Functional |
| |
[0,0,228] |
[#0000E4] |
D-GENE ORF |
| |
[128,0,128] |
[#800080] |
D-GENE Pseudogene |
| |
[255,228,0] |
[#FFE400] |
J-GENE Functional |
| |
[255,228,0] |
[#FFE400] |
J-GENE ORF |
| |
[255,106,0] |
[#FF6A00] |
J-GENE Pseudogene |
| |
[0,153,250] |
[#0099FA] |
C-GENE Functional |
| |
[0,153,250] |
[#0099FA] |
C-GENE ORF |
| |
[0,153,250] |
[#0099FA] |
C-GENE Pseudogene |
| |
[228,0,228] |
[#E400E4] |
Genes not related Functional |
| |
[228,0,228] |
[#E400E4] |
Genes not related Pseudogene |
| |
|
|
Conventional VPREB and IGLL genes are represented using the same color menu as V-GENE and C-GENE, respectively. |
Color menu for regions and domains
| |
[255,255,0] |
[#FFFF00] |
L-REGION |
| |
[0,238,0] |
[#00EE00] |
V-REGION |
| |
[255,51,0] |
[#FF3300] |
D-REGION, (N-D)-REGION |
| |
[255,204,0] |
[#FFCC00] |
J-REGION |
| |
[193,217,249] |
[#C1D9F9] |
C-DOMAIN |
| |
[193,217,249] |
[#C1D9F9] |
C-REGION |
| |
[193,217,249] |
[#C1D9F9] |
CH1 |
| |
[0,117,255] |
[#0075FF] |
HINGE-REGION |
| |
[0,117,255] |
[#0075FF] |
H, H1, H2, H3, H4, H5 |
| |
[224,242,245] |
[#E0F2F5] |
CH2 |
| |
[193,217,249] |
[#C1D9F9] |
CH3, CH5, CH7 |
| |
[224,242,245] |
[#E0F2F5] |
CH4, CH6 |
| |
[0,153,0] |
[#009900] |
CHS |
| |
[193,217,249] |
[#C1D9F9] |
CL |
| |
[254,234,200] |
[#FEEAC8] |
G-DOMAIN |
| |
[254,234,200] |
[#FEEAC8] |
G-ALPHA, G-ALPHA1, G-ALPHA2, G-BETA |
| |
[161,238,161] |
[#A1EEA1] |
V-LIKE-DOMAIN |
| |
[190,228,234] |
[#BEE4EA] |
C-LIKE-DOMAIN |
| |
[254,234,200] |
[#FEEAC8] |
G-LIKE-DOMAIN, G-ALPHA1-LIKE, G-ALPHA2-LIKE |
| |
[255,204,150] |
[#FFCC96] |
CONNECTING-REGION |
| |
[212,202,217] |
[#D4CAD9] |
TRANSMEMBRANE-REGION |
| |
[255,204,204] |
[#FFCCCC] |
CYTOPLASMIC-REGION |
| |
[255,255,223] |
[#FFFFDF] |
FIBRONECTIN-DOMAIN |
Color menu for mutations
Color menu for mutations is according to Rules for description of genes and alleles in IMGT Repertoire (IMGT annotation rules)
| n |
[255,0,0] |
[#FF0000] |
nucleotide mutations and amino acid changes for a given codon |
| n |
[0,0,0] |
[#000000] |
nucleotide mutations and amino acid changes at the 3' end of V-REGION, 5' end of J-REGION, or on either end of D-REGION, which may belong to the N-REGION |
Color menu for sequence alignments
The color menu for sequence alignments is used in:
| |
[255,204,150] |
[#FFCC96] |
CONNECTING-REGION |
| |
[212,202,217] |
[#D4CAD9] |
TRANSMEMBRANE-REGION |
| |
[255,204,204] |
[#FFCCCC] |
CYTOPLASMIC-REGION |
| BC |
[128,0,0] |
[#800000] |
BC, C' and C'' loops in V-REGION, BC and FG loops in C-REGION |
| AB |
[0,0,0] |
[#000000] |
AB, CD, DE and EF turns in C-REGION |
| C |
[204,51,204] |
[#CC33CC] |
1st-CYS and 2nd-CYS (intrachain disulfide bridge) |
| W |
[0,0,255] |
[#0000FF] |
CONSERVED-TRP |
| L |
[51,102,255] |
[#3366FF] |
conserved hydrophobic amino acids (A, C, F, I, L, M, V, W) |
| N |
[0,255,0] |
[#00FF00] |
N (Asn, asparagine) is potential N-glycosylation site that belong to the N-glycosylation motif NXS/T, where X is any amino acid except proline (P) |
| M |
[255,0,0] |
[#FF0000] |
INIT-CODON (atg) |
| * |
[255,0,0] |
[#FF0000] |
STOP-CODON (taa, tga, tag) |
Color menu for splicing types
Color menu for splicing types identify the nucleotides and amino acids resulting from
the splicing (splicing frames 1 and 2) or next to the splicing (splicing frame 0).
The last nucleotide of the upstream 5' exon is shown in bold and is colored in purple (splicing frame 1),
green (splicing frame 2) or blue (splicing frame 0).
The first amino acid the downstream (3') exon is in bold and is colored in purple (splicing frame 1),
green (splicing frame 2) or blue (splicing frame 0).
For the splicing frames 1 and 2, that amino acid results from the splicing.
Each splicing type is illustrated with an example, below.
| Splicing frame 0 (codon_start1) |
 |
| N |
[50,50,205] |
[#3232CD] |
codon_start1 |
| |
[255,204,204] |
[#FFCCCC] |
INTRACYTOPLAMIC-REGION |
| Splicing frame 1 (codon_start3) |
 |
| G |
[204,51,204] |
[#CC33CC] |
codon_start3 |
| |
[255,255,0] |
[#FFFF00] |
L-REGION |
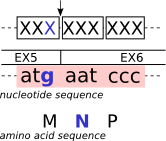
| Splicing frame 2 (codon_start2) |
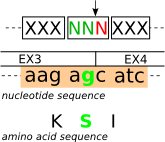 |
| S |
[0,255,0] |
[#00FF00] |
codon_start2 |
| |
[255,204,150] |
[#FFCC96] |
CONNECTING-REGION |
Color menu for splicing representation
| Spliced intron |
| |
[204,51,204] |
[#CC33CC] |
Splicing frame 1 |
| |
[0,255,0] |
[#00FF00] |
Splicing frame 2 |
| |
[50,50,205] |
[#3232CD] |
Splicing frame 0 |
| Exon or intron not included in transcript |
| |
[103,103,103] |
[#676767] |
|
| Exon or intron modified length due to an alternative splicing |
| |
[255,10,22] |
[#FF0A16] |
|
Color menu for IMGT pMH contact sites
Color menu for IMGT pMH contact sites is according to [3]
| |
[255,0,0] |
[#FF0000] |
C1 |
| |
[128,128,128] |
[#808080] |
C2 |
| |
[0,0,255] |
[#0000FF] |
C3 |
| |
[0,255,0] |
[#00FF00] |
C4 |
| |
[0,128,255] |
[#0080FF] |
C5 |
| |
[255,255,0] |
[#FFFF00] |
C6 |
| |
[153,255,51] |
[#99FF33] |
C7 |
| |
[0,255,255] |
[#00FFFF] |
C8 |
| |
[128,128,255] |
[#8080FF] |
C9 |
| |
[255,179,0] |
[#FFB300] |
C10 |
| |
[255,128,255] |
[#FF80FF] |
C11 |
Color menu for rearrangenment
| |
[64,64,64] |
[#404040] |
Productive rearrangenment |
| |
[64,64,64] |
[#404040] |
Unproductive rearrangenment |