Immunoglobulin (IG) or antibody glycosylation
Glycosylation sites
N (Asn, asparagine) are potential N-glycosylation sites when they are part of the N-glycosylation motif NXS/T, where X is any amino acid except proline. Glycans are linked to the nitrogen atom N (N-linked glycosylation)[1].
S (Ser, serine) and T (Thr, threonine) are potential glycosylation sites but the motifs are not very conserved. Glycans are linked to the oxygen atom O (O-linked glycosylation).
N-glycosylation of immunoglobulins (IG) or antibodies
Glycosylated IgG
All currently approved therapeutic antibodies on the market are IgG or derivatives.
IgG contain one conserved N-linked glycosylation site in each CH2 domain of their C-REGION. This site (asparagine, Asn N84.4) is located at the DE-TURN. They may also have secondary sites in the Fab regions but those are not conserved.
The IMGT Collier de Perles of CH2-DOMAIN is represented below for human IGHG1 (1hzh in IMGT/3Dstructure-DB) on one layer (left panel) or on two layers (right panel).
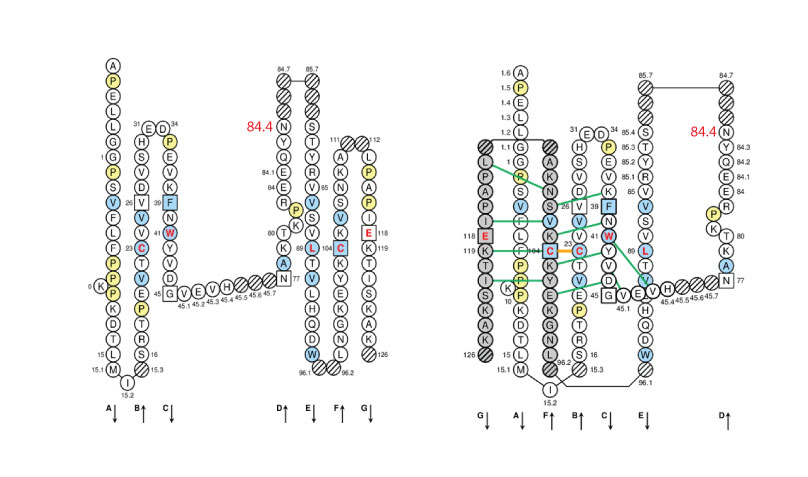
Positions at which hydrophobic amino acids are found in more than 50% of analysed IG and TR sequences are shown in blue and prolines (P) are shown in yellow. The glycosyaltion site (84.4) is indicated in red.
The 3D structure of the human IgG b12 antibody with its 2 carbohydrate chains is shown below as an example (1hzh in IMGT/3Dstructure-DB, Jmol version 12.2.15).
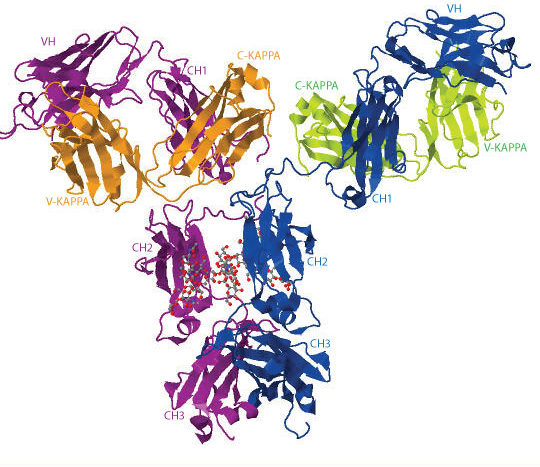
The heavy chains are represented in blue and purple, the light chains are in orange and green, respectively. The 2 N-glycans are illustrated in ball-and-stick representation.
Oligosaccharides found in human IgG
The human glycans are mainly classified as 'biantennary complex' structure with a core fucose (Fuc) and are often terminated with N-acetylneuraminic acid (Neu5Ac), a sialic acid.
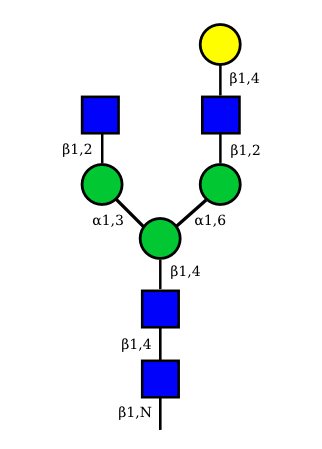
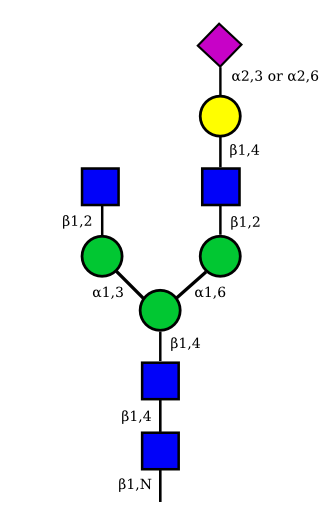
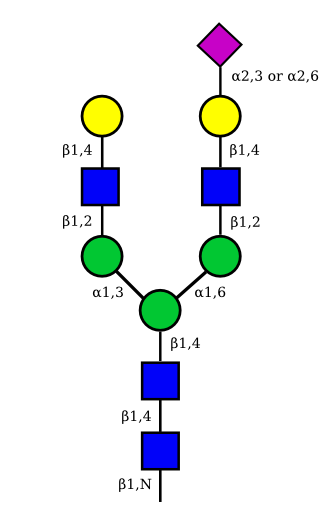
The figure below shows the largest N-linked oligosaccharide structure found in human IgG. The third N-acetylglucosamine (GlcNac, NAG) bisecting arm represents around 10% of human IgGs glycoforms [2].
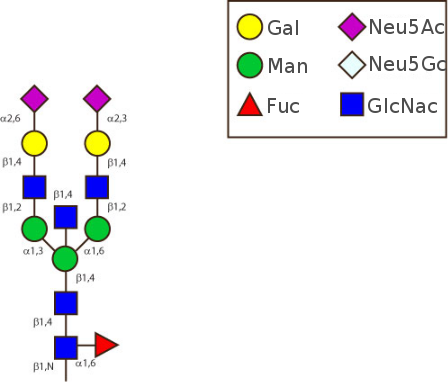
Gal: galactose, Man: mannose, Fuc: fucose, GlcNac: N-acetylglucosamine, Neu5Ac: N-acetylneuraminic acid and Neu5Gc: N-glycolylneuraminic acid. Symbol nomenclature is from the Nomenclature Committee [3-5].
Description of the carbohydrate
The 4 most abundant glycans in mAb biopharmaceuticals are shown below. The Fc oligosaccharides are terminated by 0, 1 or 2 galactoses and are called G0, G1 or G2 respectively.
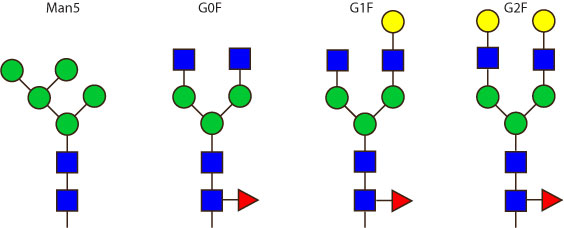
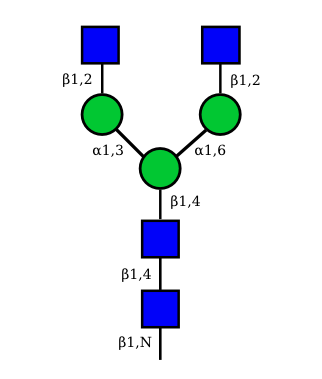
The conserved heptasaccharide core is composed of 2 N-acetylglucosamine (GlcNAc), 3 mannose (Man) and 2 other GlcNAc residues that are β-1,2 linked to α-6 Man and α-3 Man, forming two arms.
For G1F, Gal can be on the α1,3-arm or on the α1,6-arm.
Additional fucose (Fuc), galactose (Gal), N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) residues may be present or not, particularly depending on the expression system.
Current monoclonal antibodies production systems
Mammalian cell expression systems are the favorite methods for the commercial production of monoclonal antibodies because their protein glycosylation machinery closely resembles that in human.
The current marketed antibodies are mainly expressed in CHO (Chinese Hamster Ovary), SP2/0 (mouse myeloma cells), NS0 (Non-Secreting mouse myeloma cells) and hybridomas.
Glycosylation patterns of recombinant antibodies produced in human, CHO and NS0 cells
The figure bellow illustrates the main differences regarding the glycosylation patterns of antibodies produced in human cells, CHO and NS0.
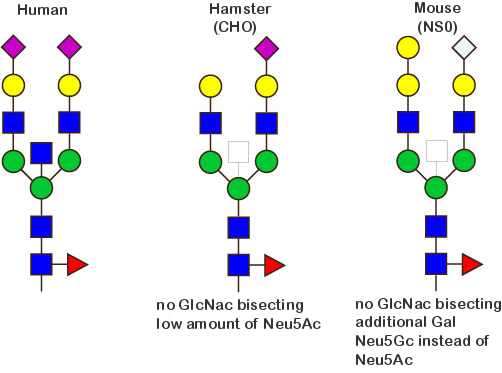
Fc glycosylation of monoclonal antibodies expressed in human cells
Monoclonal antibodies expressed in human cells are glycosylated on the IGHG CH2 N84.4 asparagin. The N-glycans are of the complex biantennary form.
There is a total of 36 possible isoforms, in 3 major classes G0, G1, G2, according to the number of terminal galactose.
| Classes | Glycan name | Nb | Fucosylated | Afucosylated | ||||
|---|---|---|---|---|---|---|---|---|
| G0 | agalactosylated | G0A0 | 4 | +B+F | -B+F | +B-F | -B-F | asialylated |
| G1 | monogalactosylated | G1A0 | 4*2 | +B+F | -B+F | +B-F | -B-F | asialylated |
| G1A1 | 4*2 | +B+F | -B+F | +B-F | -B-F | monosialylated | ||
| G2 | digalactosylated | G2A0 | 4 | +B+F | -B+F | +B-F | -B-F | asialylated |
| G2A1 | 4*2 | +B+F | -B+F | +B-F | -B-F | monosialylated | ||
| G2A2 | 4 | +B+F | -B+F | +B-F | -B-F | disialylated | ||
G0, G1, G2: with 0, 1 or 2 galactose; G0 = agalactosylated, G1 = monogalactosylated, G2 = digalactosylated
A0, A1, A2: with 0, 1 or 2 sialic acid; A0 = asialylated (or 'neutral'), A1 = monosialylated, A2 = disialylated. The percentage of A0, A1 et A2 inside each class is difficult to evaluate.
Monosialylation and monogalactosylation may occur on either the α1-3 or α1-6 arm of the biantennary structures.
+B, -B: with or without a 'bisecting' N-acetyl glucosamine, GlcNAc (between the two antennae of the glycan and attached to the position 4 of the branching mannose); +B = bisected
+F, -F: with or without a core fucose; +F = fucosylated, -F = afucosylated.
N-Glycan structures of IG expressed in human cells
The possible different structures from the table above are represented. Links when provided are to UniCarbKB.
| Glycan name | Fucosylated | Afucosylated | ||
|---|---|---|---|---|
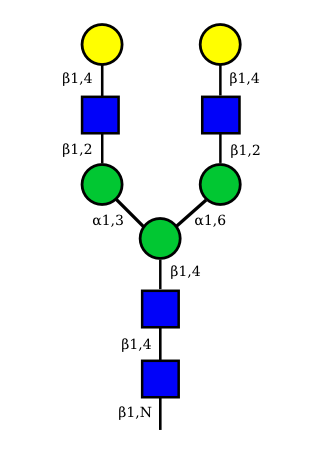
| G0A0 | 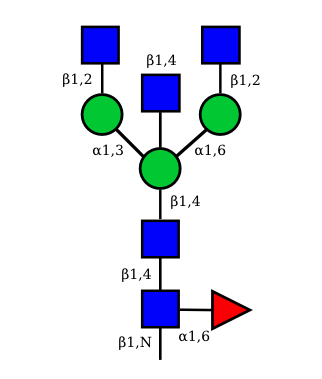 UC 1495 |
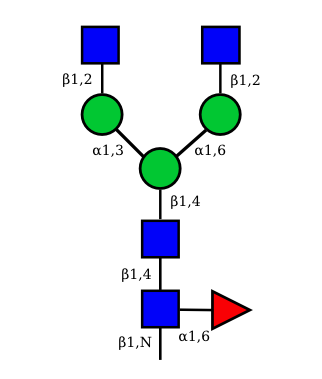 UC 1099 |
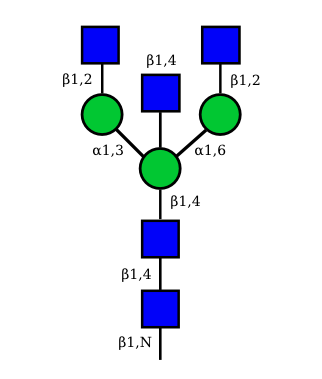 |
 UC 1623 |
| G1A0 | 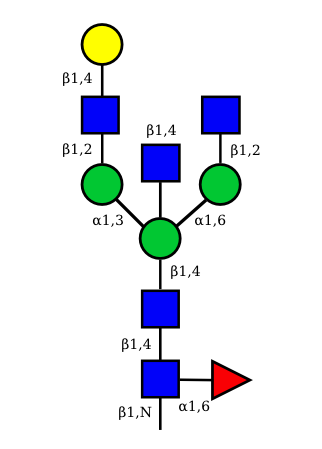 UC 1499 |
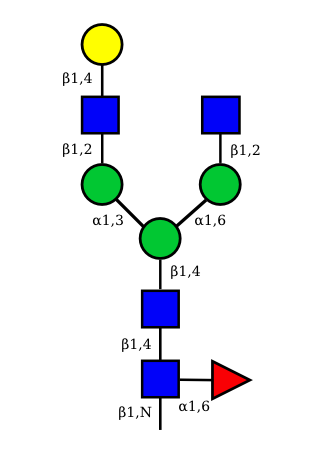 UC 1599 |
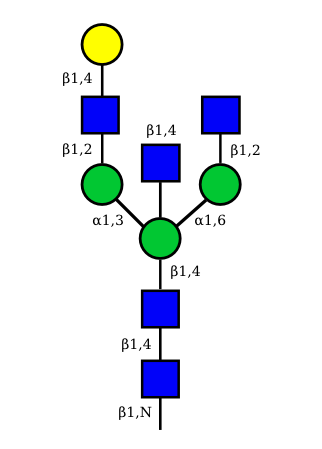 |
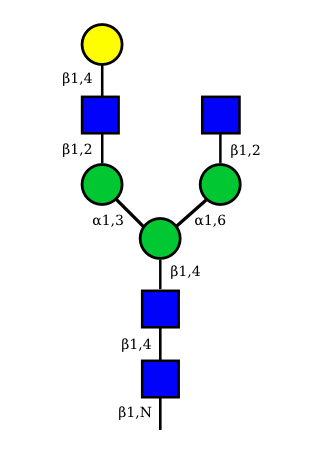 UC 1489 |
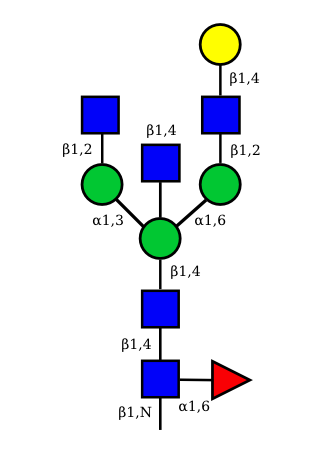 UC 1497 |
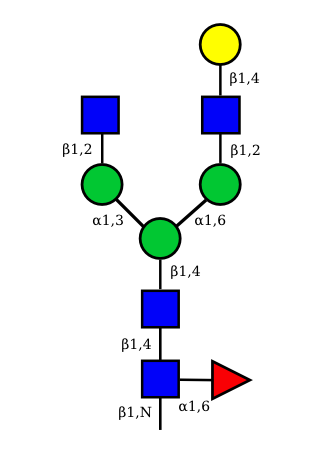 UC 1493 |
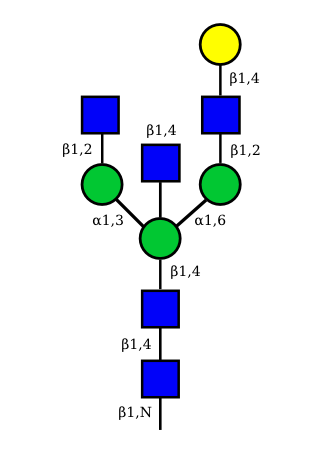 |
 UC 2073 |
|
| G1A1 | 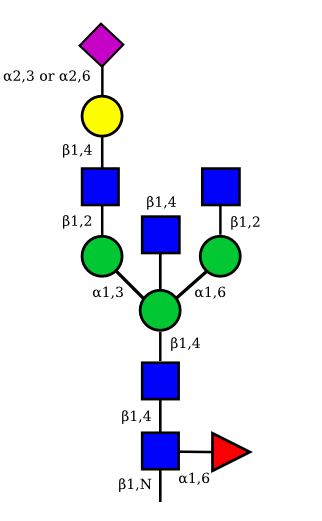 |
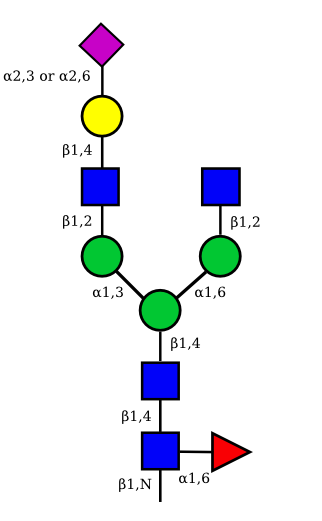 |
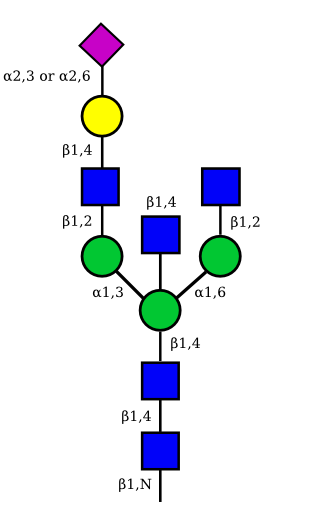 |
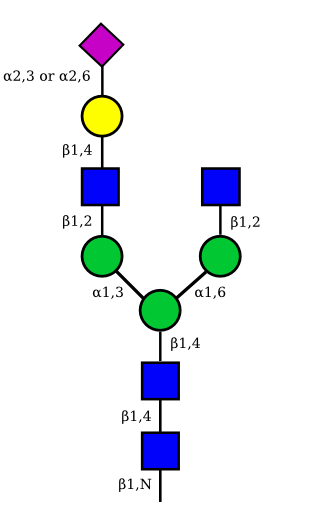 |
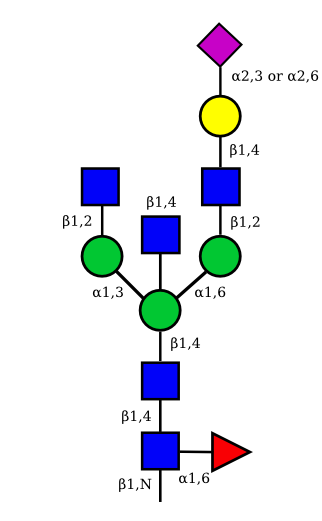 |
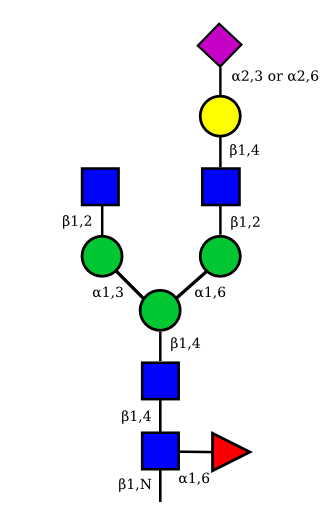 |
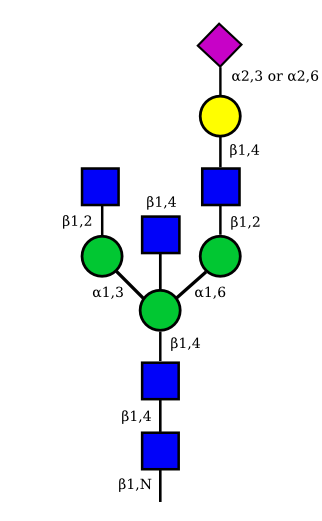 |
 |
|
| G2A0 | 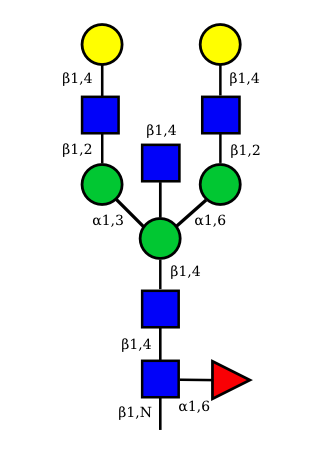 UC 1501 |
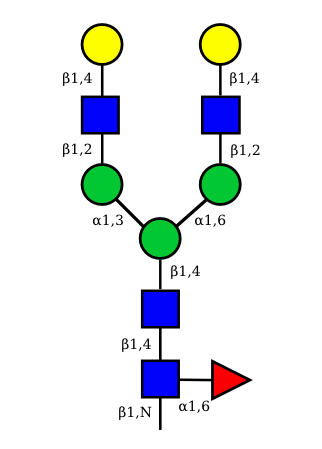 UC 1603 |
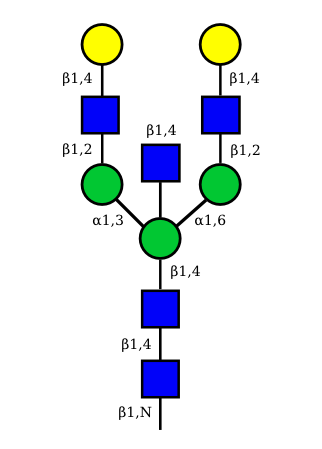 |
 UC 1521 |
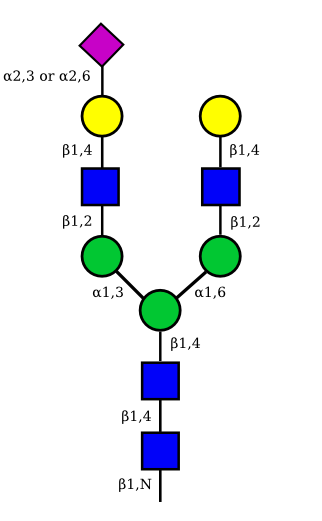
| G2A1 | 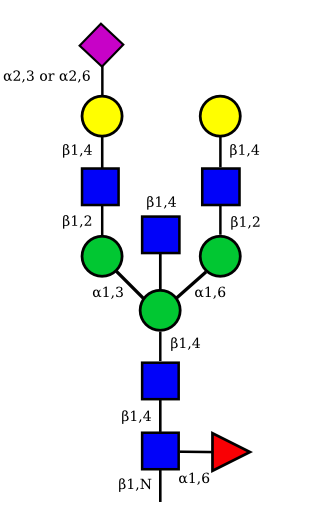 |
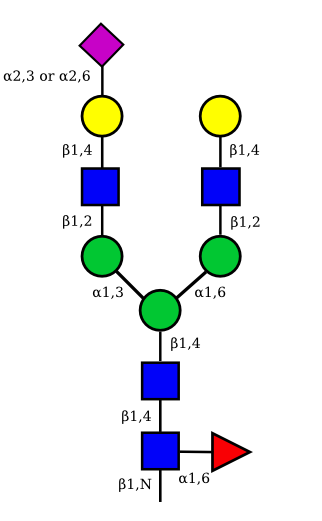 UC 1119 |
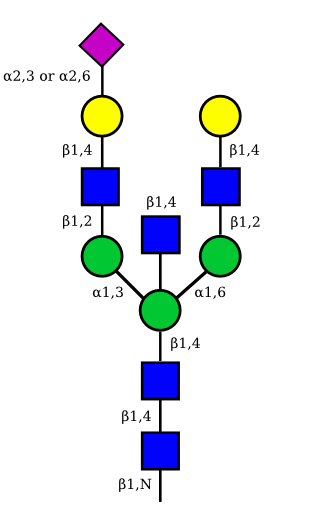 |
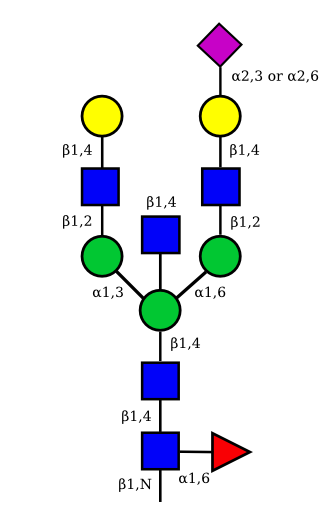
 |
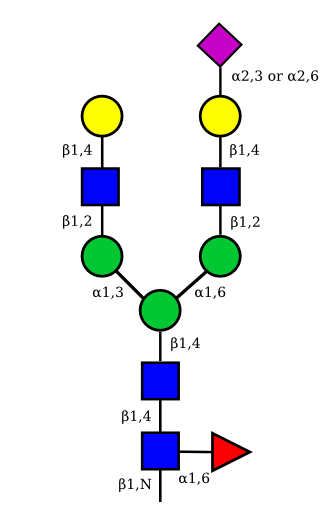
 |
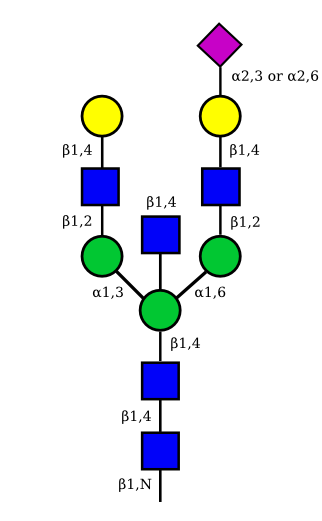
 UC 1973 |
 |
 |
|
| G2A2 | 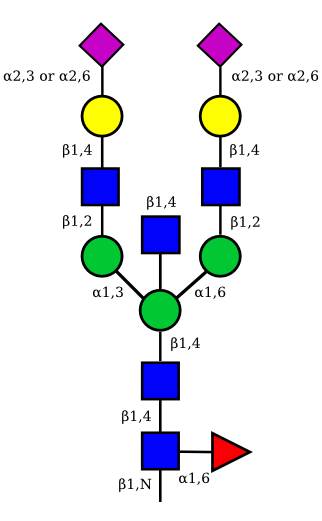 |
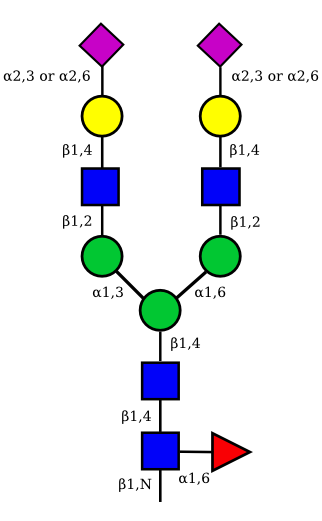 UC 1527 |
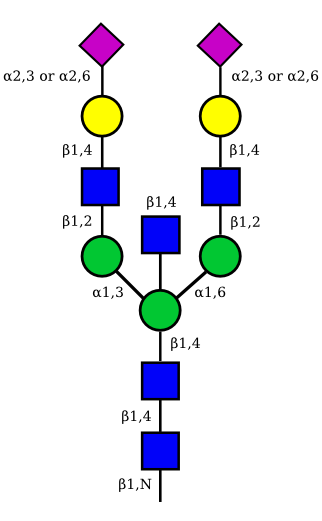 |
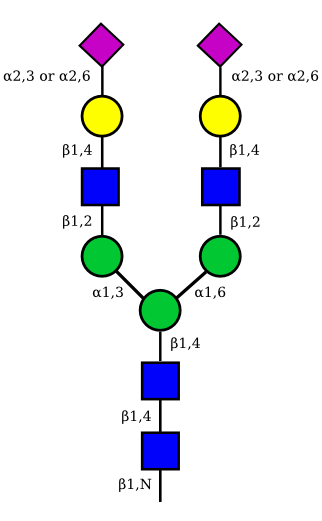 |
Other N-glycan structures
| G1F-GlcNac | G1-GlcNac |
|---|---|
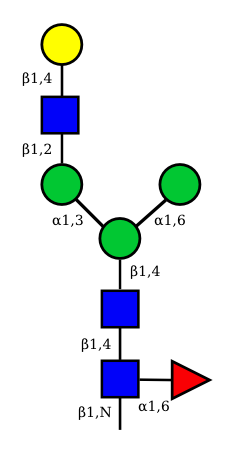 UC 2099 |
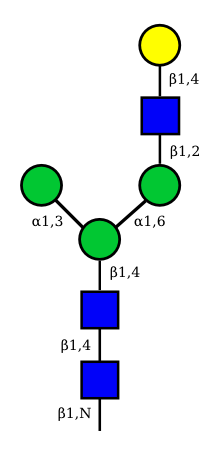
UC 3135 |
Mannose
| Man9 | Man8 | Man7 | Man6 |
|---|---|---|---|
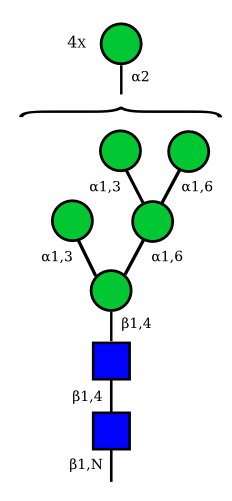 UC 1645 |
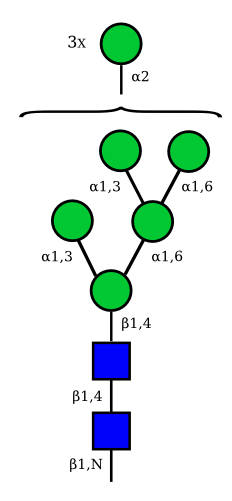
UC 1643 |
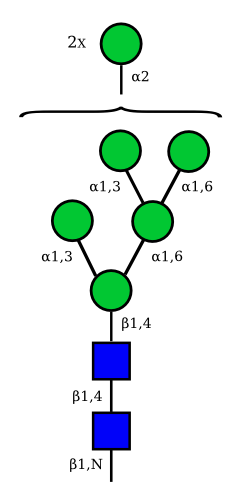
UC 1641 |
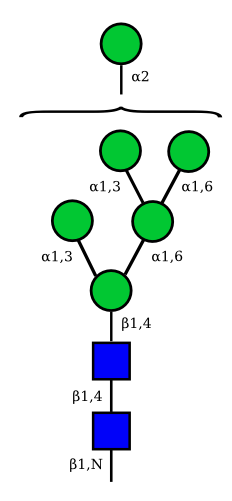
|
Correspondence between nomenclatures for the IG glycans expressed in human cells
Correspondance between the GA and HN nomenclatures is given for the 36 possible structures of IG N-glycans expressed in human cells.
The GA nomenclature is based on the number of galactose (G) (0, 1 or 2) and sialic acid (A) (0, 1 or 2) at the terminal ends of the arms of the IG N-glycans. The presence or absence of the 'bisecting' N-acetyl glucosamine (B) and of the core fucose (F) are indicated by the letter B or F preceded by a + or a -.
The HN nomenclature is based on the number of carbohydrates in the glycan composition (H: number of hexoses, N: number of N-acetyl glucosamine), with, if present, S number of sialic acids. The presence of the core fucose is indicated by the letter F. The number between square brackets represents the total number of monosaccharides in each structure.
The graphical representation of the 36 possible structures of IG N-glycans expressed in human cells are shown above.
| Glycan name subclass |
Fucosylated | Afucosylated | |||
|---|---|---|---|---|---|
| +B+F | -B+F | +B-F | -B-F | ||
| G0A0 | G0A0+B+F | G0A0-B+F | G0A0+B-F | G0A0-B-F | asialylated |
| H3N4 [7] | H3N5F [9] | H3N4F [8], G0, | H3N5 [8] | H3N4, G0(-F) | |
| G1A0 | G1A0+B+F | G1A0-B+F | G1A0+B-F | G1A0-B-F | asialylated |
| H4N4 [8] | H4N5F [10] | H4N4F [9] | H4N5 [9] | H4N4 [8] | G1 on α1-3 arm |
| H4N5F [10] | H4N4F [9] | H4N5 [9] | H4N4 [8] | G1 on α1-6 arm | |
| G1A1 | G1A1+B+F | G1A1-B+F | G1A1+B-F | G1A1-B-F | monosialylated |
| H4N4S1 [9] | H4N5S1F [11] | H4N4S1F [10] | H4N5S1 [10] | H4N4S1 [9] | G1 and A1 (S1) on α1,3 arm |
| H4N5S1F [11] | H4N4S1F [10] | H4N5S1 [10] | H4N4S1 [9] | G1 and A1 (S1) on α1,6 arm | |
| G2A0 | G2A0+B+F | G2A0-B+F | G2A0+B-F | G2A0-B-F | asialylated |
| H5N4 [9] | H5N5F [11] | H5N4F [10] | H5N5 [10] | H5N4 [9] | |
| G2A1 | G2A1+B+F | G2A1-B+F | G2A1+B-F | G2A1-B-F | monosialylated |
| H5N4S1 [10] | H5N5S1F [12] | H5N4S1F [11] | H5N5S1 [11] | H5N4S1 [10] | A1 (S1) on α1,3 arm. S1 bond may be α2,3 or α2,6 |
| H5N5S1F12] | H5N4S1F [11] | H5N5S1 [11] | H5N4S1 [10] | A1 (S1) on α1-6 arm. S1 bond may be α2,3 or α2,6 | |
| G2A2 | G2A2+B+F | G2A2-B+F | G2A2+B-F | G2A2-B-F | disialylated |
| H5N4S2 [11] | H5N5S2F [13] | H5N4S2F [12] | H5N5S2 [12] | H5N4S2 [11] | S1 and S2 bonds may be α2,3 or α2,6 |
Influence of the glycosylation modification
Glycosylation of IgG antibodies have been shown to have a large effect on the immunogeneicity, solubility and half-life.
High mannose N-glycans
| Mannose name | ||
|---|---|---|
| H4N2 | Man4N2 | 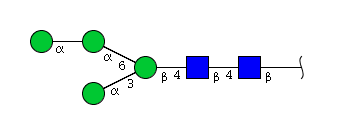 |
| H4N2 | Man4N2 | 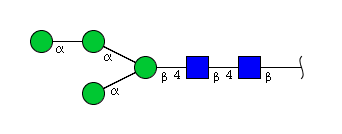 |
| H4N2 | Man4N2 | 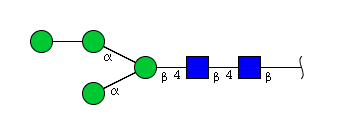 |
| H4N2 | Man4N2 |  |
| H4N2 | Man4N2 | 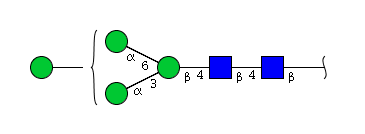 |
| H4N2 | Man4N2 | 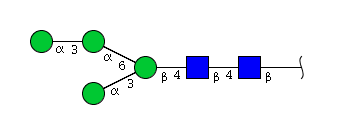 |
| H4N2 | Man4N2 | 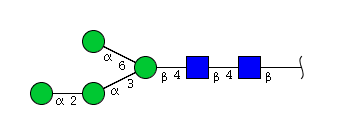 |
| H4N2 | Man4N2 | 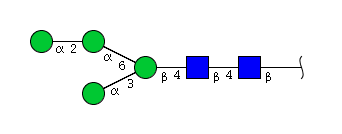 |
| H4N2 | Man4N2 | 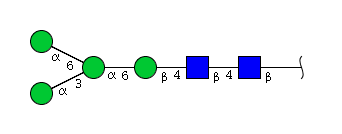 |
| H4N2 | Man4N2 | 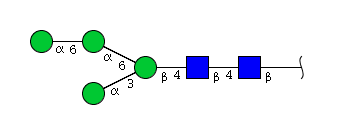 |
| H4N2 | Man3N2G1 | 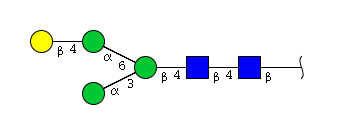 |
| H4N2 | Man4N2 | 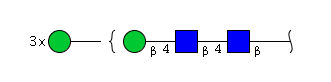 |
| H4N2 | Man4N2 |  |
| H4N2 | Man2N2G2 |  |
| H5N2 | Man5N2 | 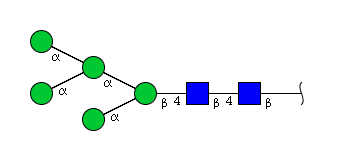 |
| H5N2 | Man5N2 | 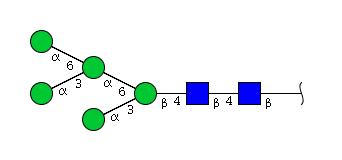 |
| H5N2 | Man5N2 | 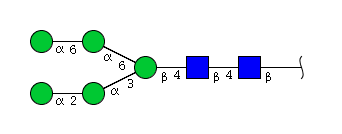 |
| H5N2 | Man5N2 | 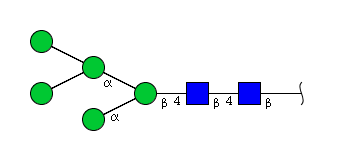 |
| H5N2 | Man5N2 | 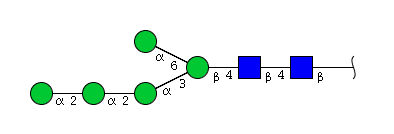 |
| H5N2 | Man5N2 |  |
| H5N2 | Man4N2G1 |  |
| H5N2 | Man5N2 |  |
| H5N2 | Man4N2G1 | 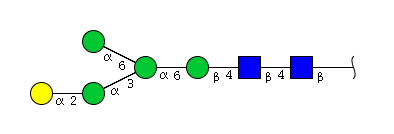 |
| H5N2 | Man5N2 | 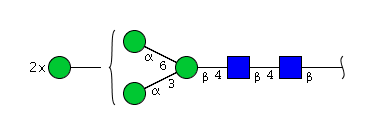 |
| H5N2 | Man5N2 | 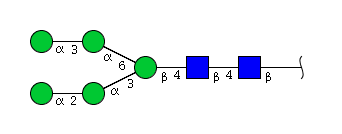 |
| H4N3 | Man3N3G1 | 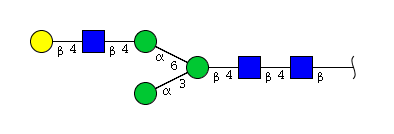 |
| H4N3 | Man3N3G1 | 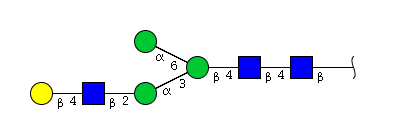 |
| H4N3 | Man3N3G1 | 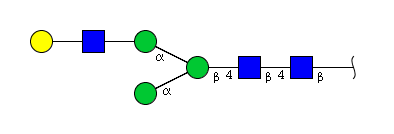 |
| H4N3 | Man4N3 | 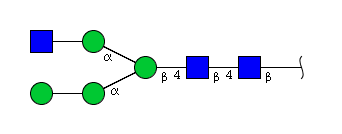 |
| H4N3 | Man3N3G1 | 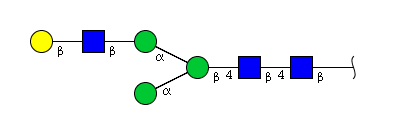 |
| H4N3 | Man4N3 | 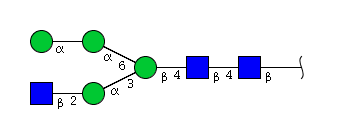 |
| H4N3 | Man3N3G1 | 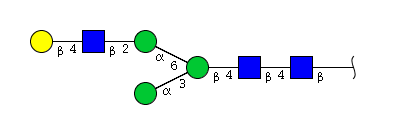 |
| H4N3 | Man4N3 | 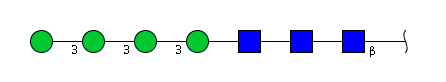 |
| H4N3 | Man4N3 | 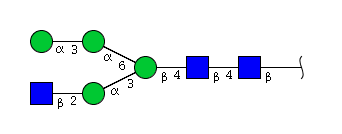 |
| H4N3 | Man2N3G2 | 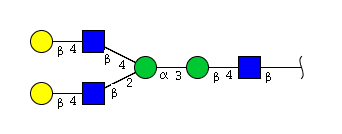 |
| H4N3 | Man3N3G1 | 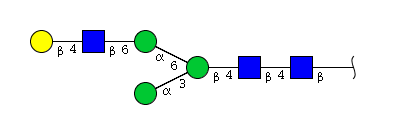 |
| H4N3 | Man3N3G1 | 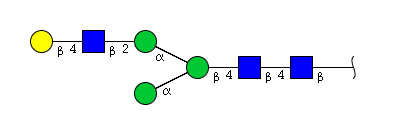 |
| H4N3 | Man4N3 | 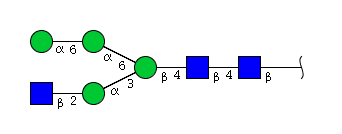 |
| H4N3 | Man3N3G1 | 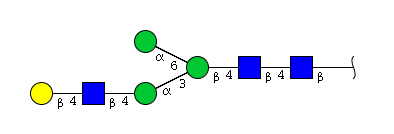 |
| H4N4 | Man3N4G1 | 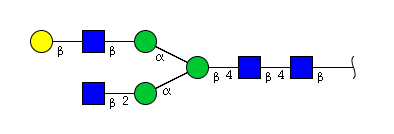 |
| H4N4 | Man3N4G1 | 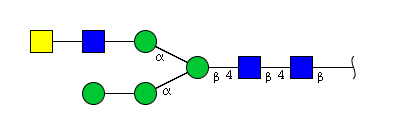 |
| H4N4 | Man3N4G1 | 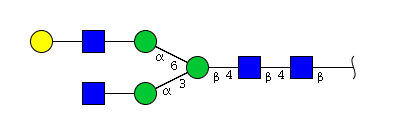 |
| H4N4 | Man3N4G1 | 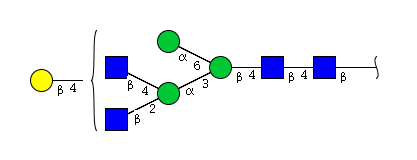 |
| H4N4 | Man3N4G1 | 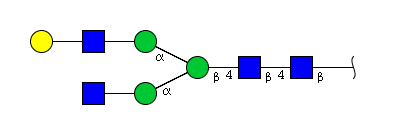 |
| H4N4 | Man3N4G1 | 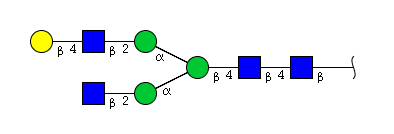 |
| H4N4 | Man4N4 | 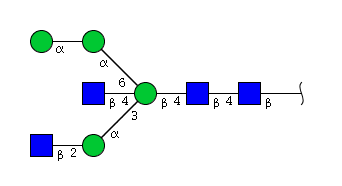 |
| H4N4 | Man4N4 |  |
| H4N4 | Man3N4G1 |  |
| H4N4 | Man3N4G1 |  |
| H4N4 | Man3N4G1 |  |
| H4N4 | Man3N4G1 | 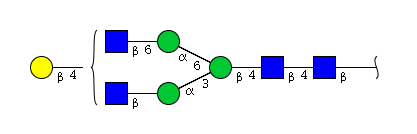 |
| H4N4 | Man3N4G1 | 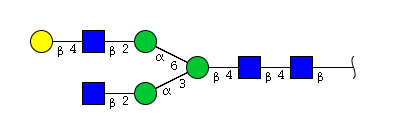 |
| H4N4 | Man3N4G1 | 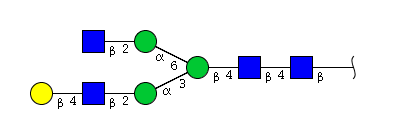 |
| H6N2 | Man6N2 | 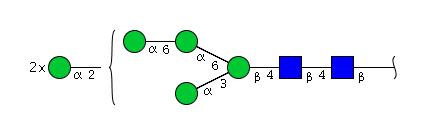 |
| H6N2 | Man6N2 | 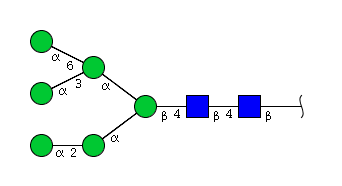 |
| H6N2 | Man6N2 | 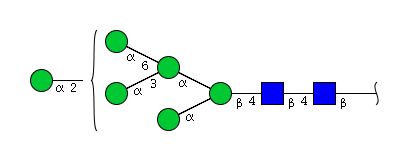 |
| H6N2 | Man6N2 | 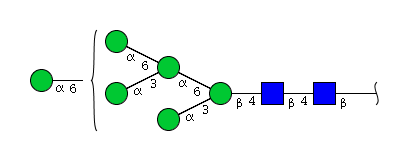 |
| H6N2 | Man6N2 | 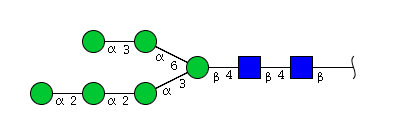 |
| H6N2 | Man6N2 | 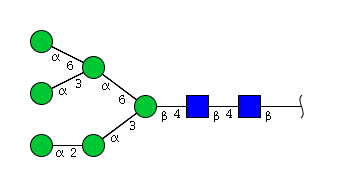 |
| H6N2 | Man6N2 | 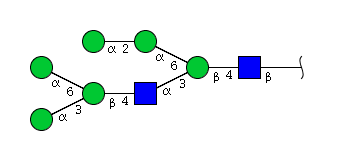 |
| H6N2 | Man6N2 | 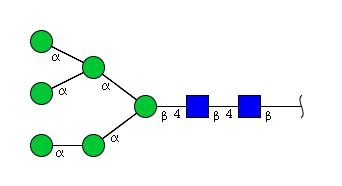 |
| H6N2 | Man6N2 | 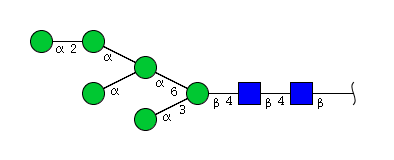 |
| H6N2 | Man6N2 | 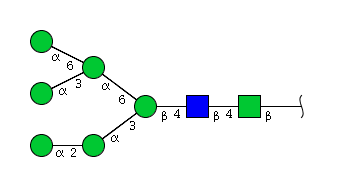 |
| H6N2 | Man6N2 | 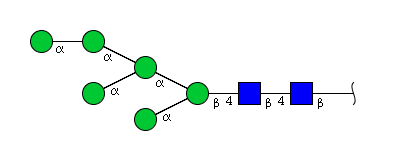 |
| H6N2 | Man6N2 | 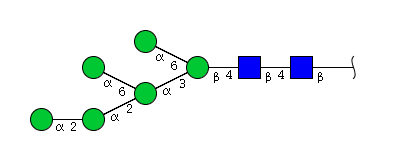 |
| H6N2 | Man6N2 | 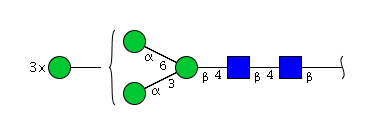 |
| H6N2 | Man6N2 | 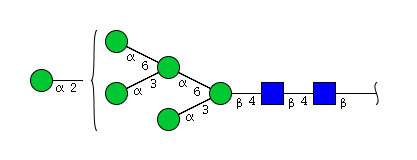 |
| H6N2 | Man6N2 | 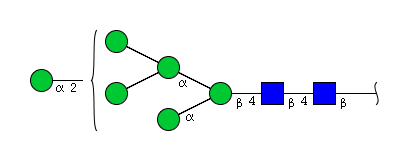 |
| H6N2 | Man6N2 | 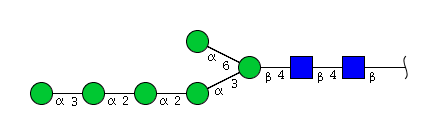 |
| H6N2 | Man6N2 | 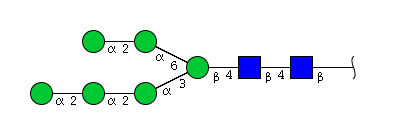 |
| H6N2 | Man5N2G1 | 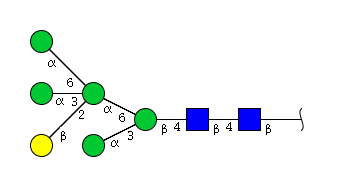 |
| H6N2 | Man6N2 | 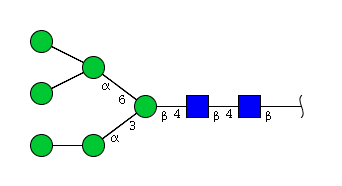 |
| H6N2 | Man5N2Glu1 | 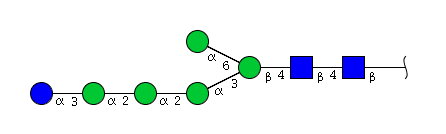 |
| H6N2 | Man5N2G1 | 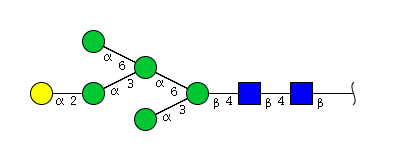 |
| H6N2 | Man6N2 | 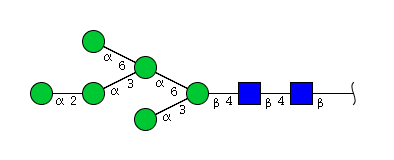 |
| H6N2 | Man6N2 | 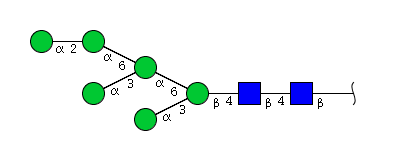 |
| H5N3 | Man3N3G2 | 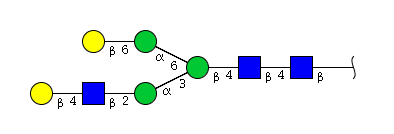 |
| H5N3 | Man3N3G2 | 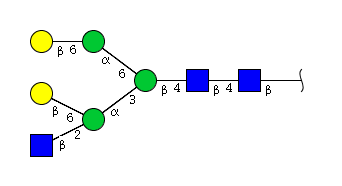 |
| H5N3 | Man4N3G1 | 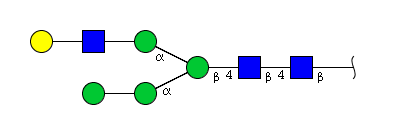 |
| H5N3 | Man4N3G1 | 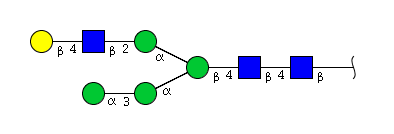 |
| H5N3 | Man5N3 | 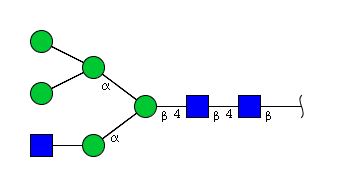 |
| H5N3 | Man4N3G1 | 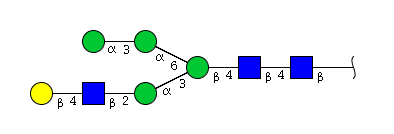 |
| H5N3 | Man5N3 | 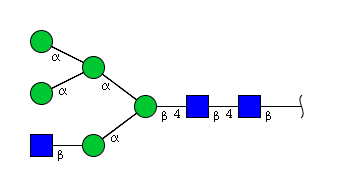 |
| H5N3 | Man5N3 | 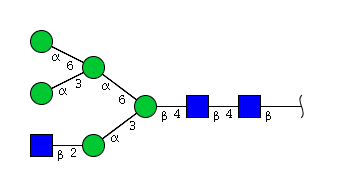 |
| H7N2 | Man7N2 | 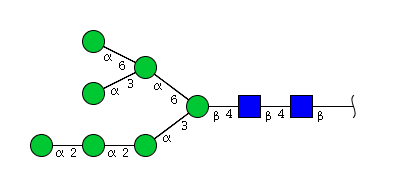 |
| H7N2 | Man5N2Glu2 | 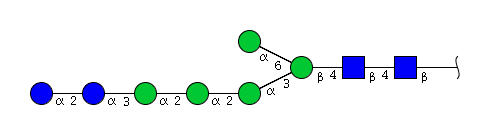 |
| H7N2 | Man7N2 | 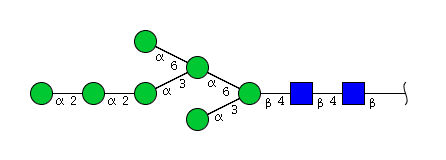 |
| H7N2 | Man7N2 | 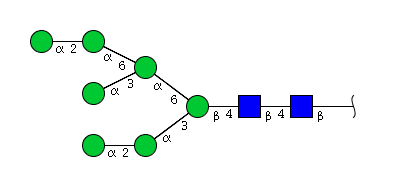 |
| H7N2 | Man7N2 | 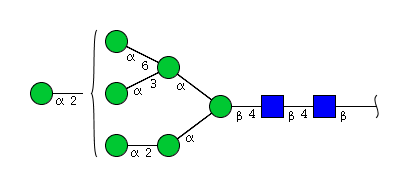 |
| H7N2 | Man7N2 | 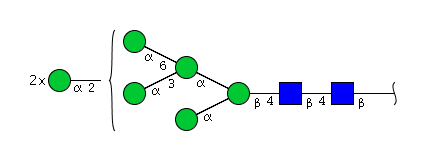 |
| H7N2 | Man7N2 | 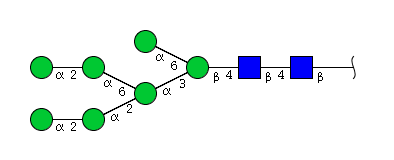 |
| H7N2 | Man7N2 | 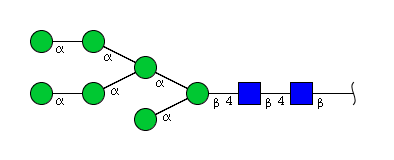 |
| H7N2 | Man6N2G1 | 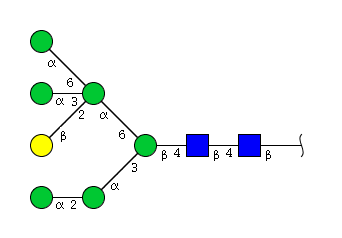 |
| H7N2 | Man7N2 | 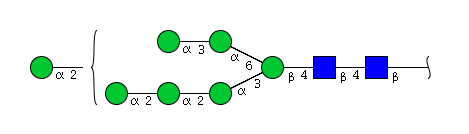 |
| H7N2 | Man6N2Glu1 | 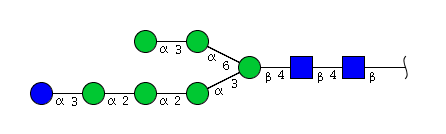 |
| H7N2 | Man7N2 | 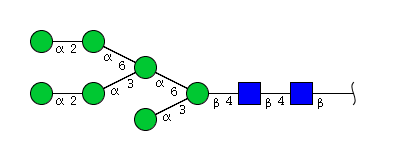 |
| H7N2 | Man7N2 | 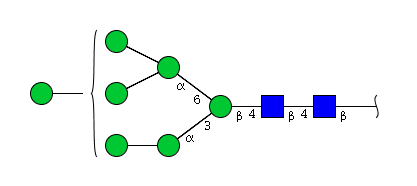 |
| H7N2 | Man7N2 | 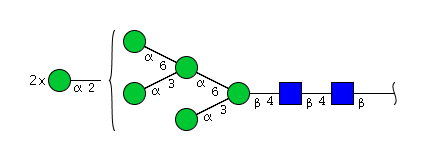 |
| H7N2 | Man5N2Glu2 |  |
| H7N2 | Man7N2 |  |
| H7N2 | Man7N2 |  |
| H7N2 | Man7N2 |  |
| H7N2 | Man7N2 | 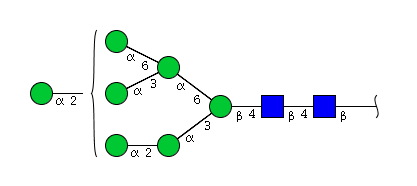 |
| H7N2 | Man7N2 | 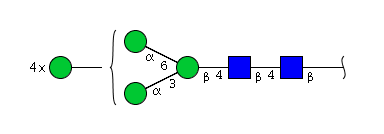 |
| H7N2 | Man7N2 | 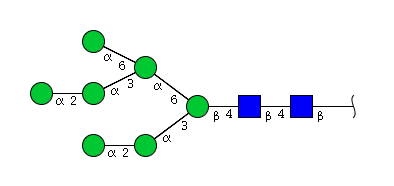 |
| H7N2 | Man7N2 | 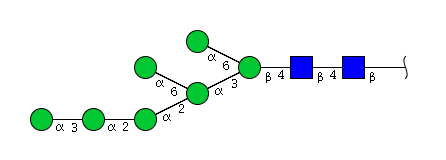 |
| H7N2 | Man7N2 | 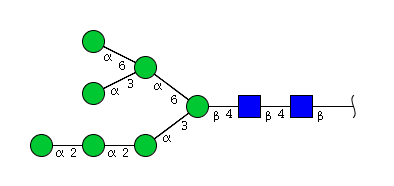 |
| H7N2 | Man6N2G1 | 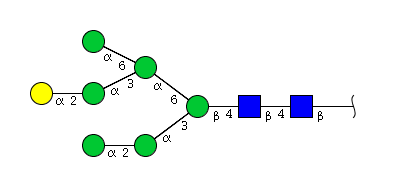 |
| H6N3 | Man5N3G1 | 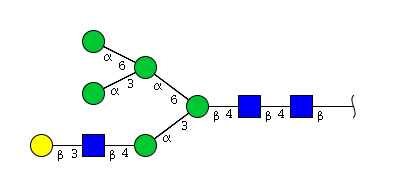 |
| H6N3 | Man5N3G1 | 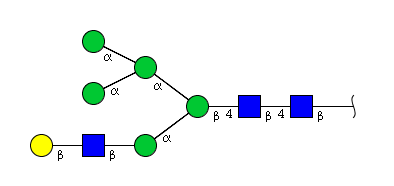 |
| H6N3 | Man5N3G1 | 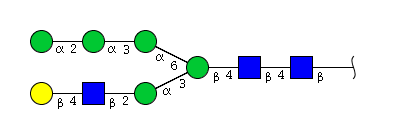 |
| H6N3 | Man5N3G1 | 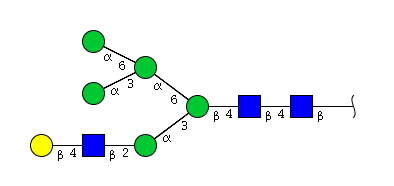 |
| H6N3 | Man5N3G1 | 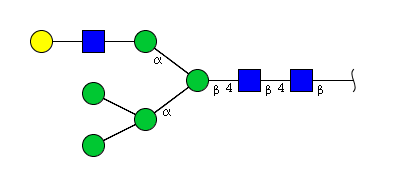 |
| H6N3 | Man5N3G1 | 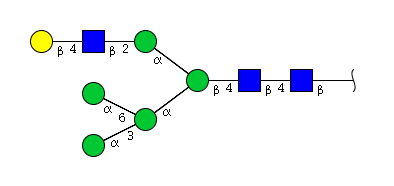 |
| H6N3 | Man6N3 | 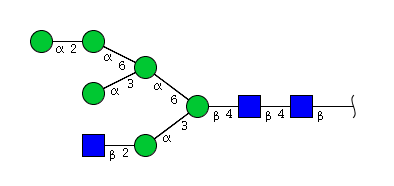 |
| H8N2 | Man7N2G1 | 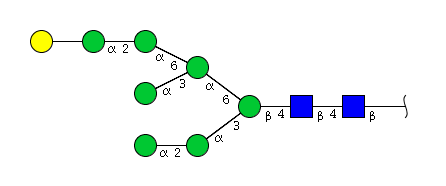 |
| H8N2 | Man8N2 | 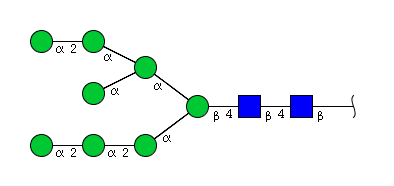 |
| H8N2 | Man8N2 | 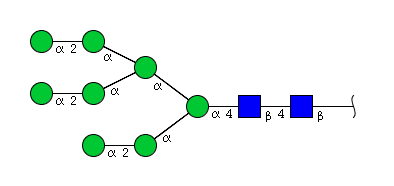 |
| H8N2 | Man8N2 | 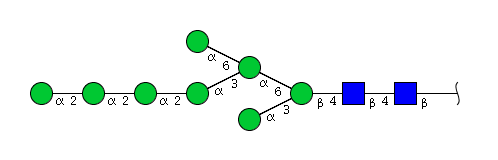 |
| H8N2 | Man7N2G1 | 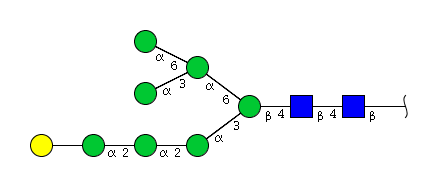 |
| H8N2 | Man8N2 | 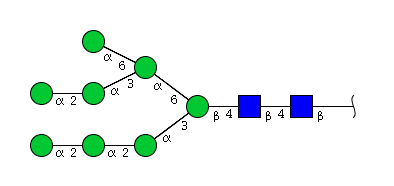 |
| H8N2 | Man5N2Glu3 | 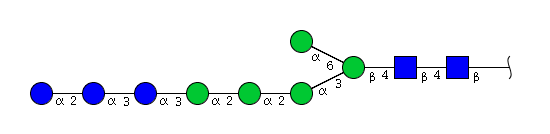 |
| H8N2 | Man7N2G1 | 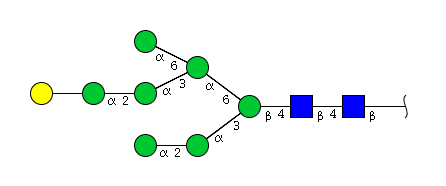 |
| H8N2 | Man8N2 | 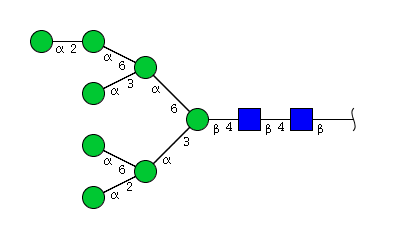 |
| H8N2 | Man7N2Glu1 | 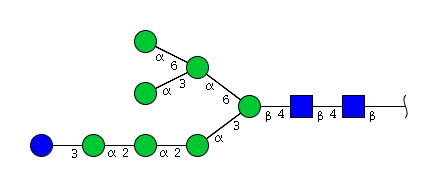 |
| H8N2 | Man8N2 | 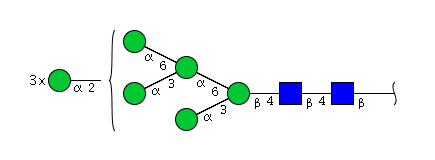 |
| H8N2 | Man8N2 | 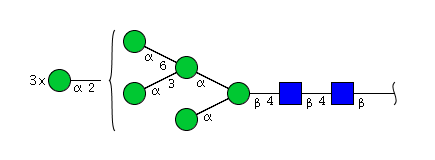 |
| H8N2 | Man8N2 | 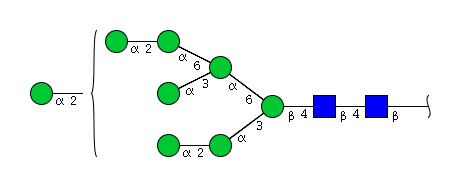 |
| H8N2 | Man8N2 | 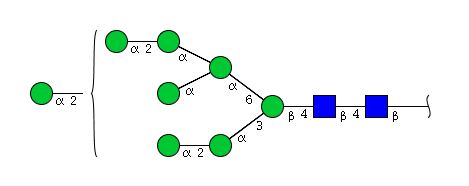 |
| H8N2 | Man8N2 | 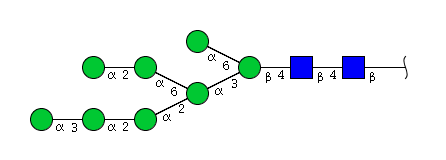 |
| H8N2 | Man7N2Glu1 | 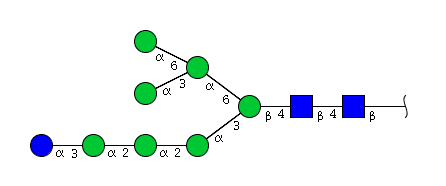 |
| H8N2 | Man7N2Glu1 | 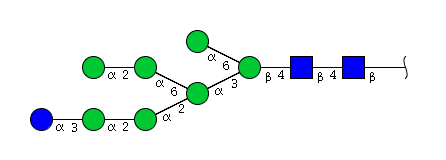 |
| H8N2 | Man7N2G1 | 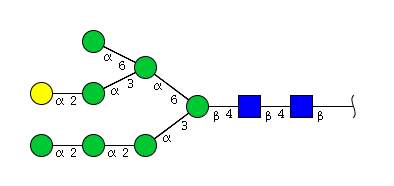 |
| H8N2 | Man8N2 | 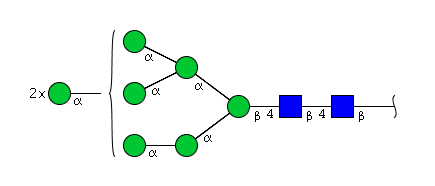 |
| H8N2 | Man8N2 | 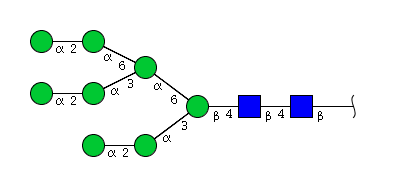 |
| H8N2 | Man8N2 | 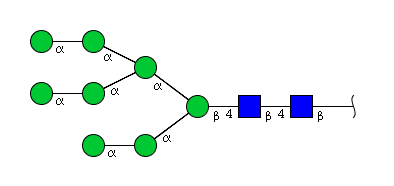 |
| H8N2 | Man7N2G1 | 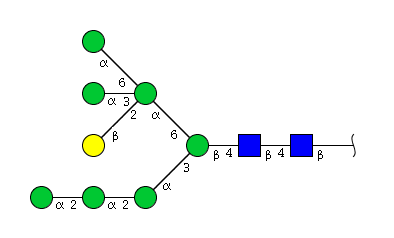 |
| H8N2 | Man8N2 | 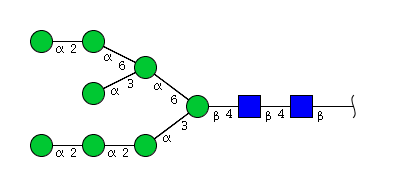 |
| H8N2 | Man8N2 |  |
| H8N2 | Man8N2 |  |
| H8N2 | Man8N2 |  |
| H8N2 | Man8N2 |  |
| H8N2 | Man7N2G1 | 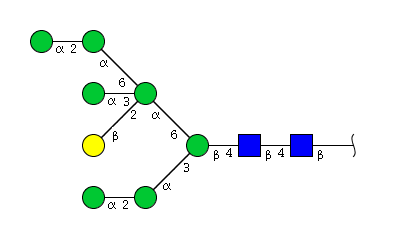 |
| H7N2 | Man7N2 |  |
| H7N2 | Man5N2Glu |  |
| H9N2 | Man8N2G1 | 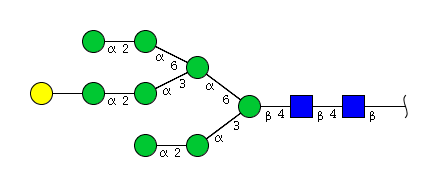 |
| H9N2 | Man8N2G1 | 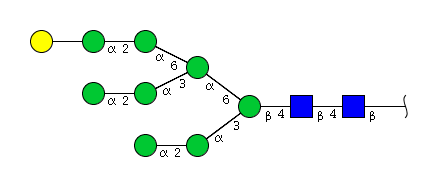 |
| H9N2 | Man7N2Glu2 | 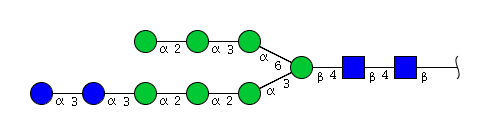 |
| H9N2 | Man9N2 | 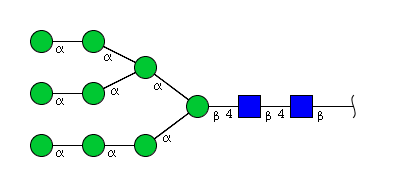 |
| H9N2 | Man9N2 | 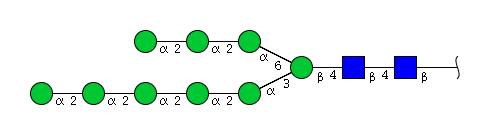 |
| H9N2 | Man8N2G1 | 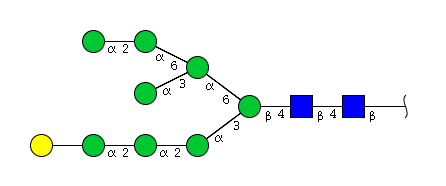 |
| H9N2 | Man8N2G1 | 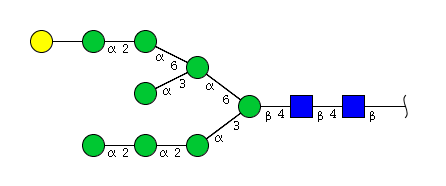 |
| H9N2 | Man9N2 |  |
| H9N2 | Man9N2 |  |
| H9N2 | Man8N2Glu1 |  |
| H9N2 | Man9N2 |  |
| H9N2 | Man8N2Glu1 | 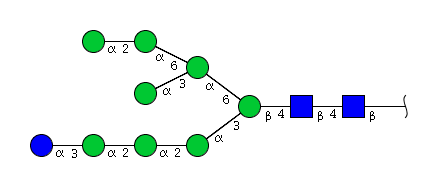 |
| H9N2 | Man8N2Glu1 | 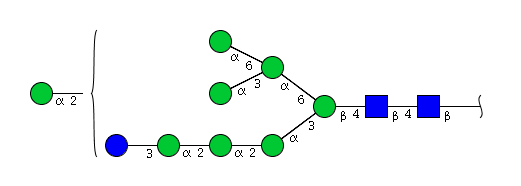 |
| H9N2 | Man8N2Glu1 | 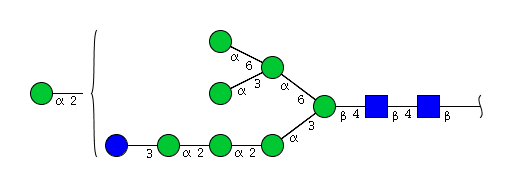 |
| H9N2 | Man8N2Glu1 | 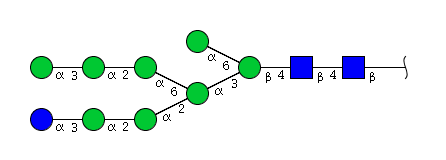 |
| H9N2 | Man9N2 | 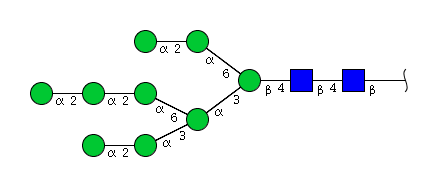 |
| H9N2 | Man7N2Glu2 | 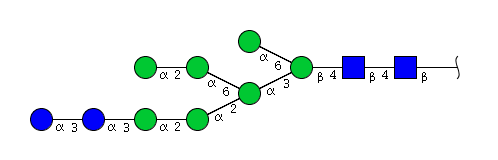 |
| H9N2 | Man8N2G1 | 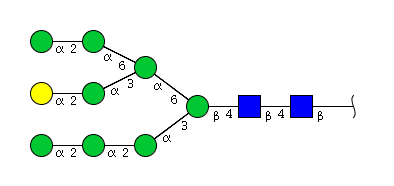 |
| H9N2 | Man8N2Glu1 | 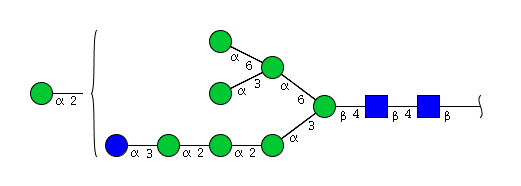 |
| H9N2 | Man9N2 | 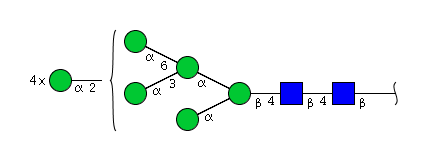 |
| H9N2 | Man9N2 | 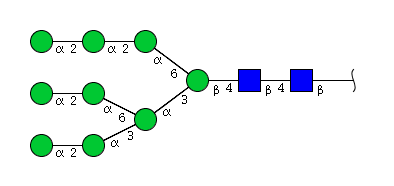 |
| H9N2 | Man9N2 | 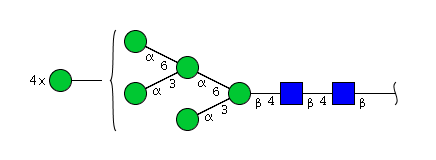 |
| H9N2 | Man9N2 | 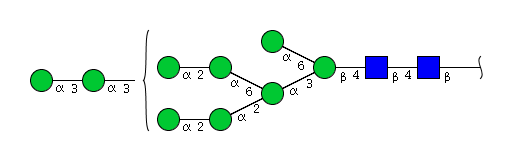 |
| H9N2 | Man9N2 | 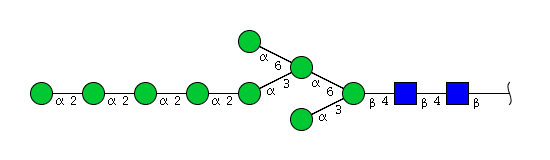 |
| H9N2 | Man9N2 | 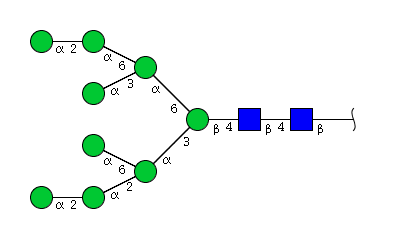 |
| H9N2 | Man9N2 | 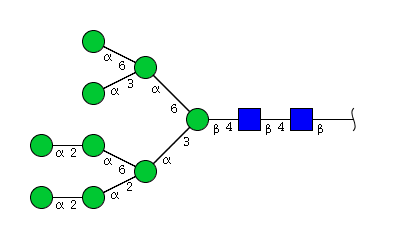 |
| H9N2 | Man9N2 | 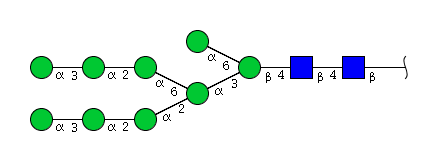 |
| H9N2 | Man9N2 | 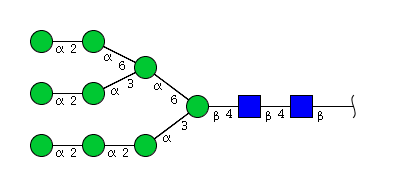 |
| H9N2 | Man9N2 | 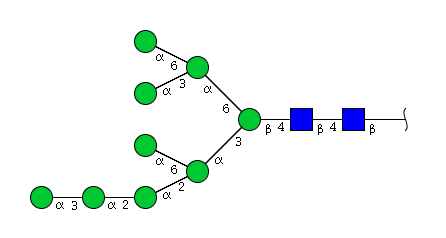 |
- Sialic acids (IMGT Lexique)
- Structural and biological properties of human immunoglobulins (IMGT Education)
- ADCC and CDC (IMGT Lexique)
| [1] | Mellquist, J.L. et al. (1998) The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37, 6833-6837. PMID: 9578569 |
| [2] | Raju, T.S. (2003) Glycosylation variations with expression systems and their impact on biological activity of therapeutic immunoglobulins. BioProcess International, 1(4), 44-53. |
| [3] | Varki, A. . et al. (2009) Symbol nomenclature for glycan representation. Proteomics. 20099(24): 5398-5399. PMID: 19902428 |
| [4] | Varki, A. et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology, 25(12):1323-1324. PMID: 26543186 |
| [5] | Appendix 1B. Symbol Nomenclature for Glycans (SNFG). In Essentials of Glycobiology [Internet]. 3rd edition (2015). Cold Spring Harbor Laboratory Press (NY). |
| Glycomics. Databases and tools at Expasy. NCBI Glycans website https://www.ncbi.nlm.nih.gov/glycans/ |



