


© Copyright 1995-2026 IMGT®, the international ImMunoGeneTics information system® | Terms of use | About us | Contact us | Citing IMGT
The IGHC protein display numbering is according to the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN.
Letters in red correspond to amino acids which are polymorphic in the other alleles.
For C-REGIONs, letters in bold correspond to additional positions in the IMGT unique numbering.
For C-REGIONs, letters between parentheses correspond to amino acids resulting from the splicing.
N (Asn, asparagine) of potential N-glycosylation sites (NXS/T, where X is different from P), (N-linked glycosylation) is shown is green (site is underlined in CHS and in pages edited before 14/10/2009).
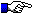 The hinge exon(s) are shown at the end of the protein display.
The hinge exon(s) are shown at the end of the protein display.
_____________________________________________________________________________________________________________________________________________________________________________________________
IGHC
genes
_____________________________________________________________________________________________________________________________________________________________________________________________
A AB B BC C CD D DE E EF F FG G CHS
--------------> ----------> ------> -------> -----------> -------> ---------->
1 10 15 16 20 23 30 36 39 41 45 77 84 85 89 96 97 104 110 118 121 130 140 150
7654321|........|....|123|...|...... ...|.....| |.....|1234567|......|12345677654321|..........|12|....... .....|....... ..|.........|.........|.........|
CH1
AJ305046,IGHE (V)SKQAPLILPLAACCKDTK...TTNITLGCLVK GYFP..EPVT VTWDAGSLNR....STMTFPAVFDQTS.....GLYTTISRVVASGKW....AKQKFTC NVVHS....QETF NKTFN
AJ302055,IGHG1 (A)STTAPKVFALAPGCGTTS...DSTVALGCLVS GYFP..EPVK VSWNSGSLTS....GVHTFPSVLQSS......GFYSLSSMVTVPASSW...TSETYIC NVVHAA..SNFKV DKRI
AJ302056,IGHG2 (A)STTAPKYFQLTPSCGITS...DATVALGCLVS DYYP..EPVT VSWNSGALTS....GVHTFPSVLQSS......GLYALSSMVTVPASTW...TSETYIC NVAHPA..SSTKV DKRI
AJ312379,IGHG3 (A)STTAPKVFPLAPSCGTTS...DSTVALGCLVS SYFP..EPVT VSWNSGTLTS....GVRTFPSVLQSS......GLYSLSSMVTVPASSL...ESKTYIC NVAHPA..SSTKV DKRI
AJ302058,IGHG4 (A)STTAPKVFPLASHSAATS...GSTVALGCLVS SYFP..EPVT VSWNSGALTS....GVHTFPSVLQSS......GLYSLSSMVTVPASSL...KSQTYIC NVAHPA..SSTKV DKKI
AJ312380,IGHG5 (E)SPKAPDVFPLTICGNTP....DPTVPVGCLVS NYFP..EPVT VSWNCDALKG....DIHTFPLDLSNS......AHHSLSSMVAVPRSS....LNQTYIC SVAHPA..SSTKV DKRI
AJ312381,IGHG6 (A)STTAPKVFQLASHSAGTS...DSTVALGCLVS SYFP..EPVT VSWNSGALTS....GVHTFPSVRQSS......GLYSLSSMVTVPASSL...KSQTYIC NVAHPA..SSTKV DKRI
L49414 ,IGHM c (E)STKTPDLFPLVSCGPSLD...ESLVAVGCLAR DFLP..NVIT FSLELPEQHCSQYPGHHEFPSVLREG.......KYSASSQVLLPSG......DVPLVC TVNHS....NGNE KVEVRPQ
CH2
AJ305046,IGHE (A)CIVTFTPPTVKLFHSSCDPGGDS.HTTIQLLCLIS DYTP..GDID IVWLIDGQKVDE..QFPQHGLVKQEG......KLASTHSELNITQGQW..ASENTYTC QVTYK....DMIF KDQARKCT
AJ302055,IGHG1 (A)PELLGGPSVFIFPPNPKDTLMI.TRTPEVTCVVV DVSQENPDVK FNWYMDGVEVR...TATTRPKEEQFN......STYRVVSVLRIQHQDW..LSGKEFKC KVNNQA..LPQPI ERTITKTK
AJ302056,IGHG2 (G)GPSVFIFPPNPKDALMI.SRTPVVTCVVV NLSDQYPDVQ FSWYVDNTEVH...SAITKQREAQFN......STYRVVSVLPIQHQDW..LSGKEFKC SVTNVG..VPQPI SRAISRGK
AJ312379,IGHG3 (A)PELLGGPSVFIFPPKPKDVLMI.TRTPEVTCLVV DVSHDSSDVL FTWYVDGTEVK...TAKTMPNEEQNN......STYRVVSVLRIQHQDW..LNGKKFKC KVNNQA..LPAPV ERTISKAT
AJ302058,IGHG4 (E)CLSVGPSVFIFPPKPKDVLMI.SRTPTVTCVVV DVGHDFPDVQ FNWYVDGVETH...TATTEPKQEQNN......STYRVVSILAIQHKDW..LSGKEFKC KVNNQA..LPAPV QKTISKPT
AJ312380,IGHG5 (A)PELPGGPSVFIFPPKPKDVLKI.SRKPEVTCVVV DLGHDDPDVQ FTWFVDGVETH...TATTEPKEEQFN......STYRVVSVLPIQHQDW..LSGKEFKC SVTNKA..LPAPV ERTTSKAK
AJ312381,IGHG6 (D)SKFLGRPSVFIFPPNPKDTLMI.SRTPEVTCVVV DVSQENPDVK FNWYVDGVEAH...TATTKAKEKQDN......STYRVVSVLPIQHQDW..RRGKEFKC KVNNRA..LPAPV ERTITKAK
L49414 ,IGHM c (V)LIQDESPNVTVFIPPRDAFTGPGQRTSRLVCQAT GFSP..KEIS VSWLRDGKPVES..GFTTEEVQPQNKES..WPVTYKVTSMLTITESDW..LNQKVFTC HVEHT....GGSF QKNVSSMCSP
CH3
AJ305046,IGHE (E)SDPRGVSVYLSPPSPLDLYV..SKSPKITCLVV DLANV.QGLS LNWSRESGEPL...QKHTLATSEQFN......KTFSVTSTLPVDTTDW..IEGETYKC TVSHPD..LPREV VRSIAKAP
AJ302055,IGHG1 (G)RSQEPQVYVLAPHPDEDS...KSKVSVTCLVK DFYP..PEIN IEWQSNGQPELET.KYSTTQAQQDSD......GSYFLYSKLSVDRNRW..QQGTTFTC GVMHEA.LHNHYT QKNVSKNP...GK
AJ302056,IGHG2 (G)PSRVPQVYVLPPHPDELA...KSKVSVTCLVK DFYP..PDIS VEWQSNRWPELEG.KYSTTPAQLDGD......GSYFLYSKLSLETSRW..QQVESFTC AVMHEA.LHNHFT KTDISESL...GK
AJ312379,IGHG3 (G)QTRVPQVYVLAPHPDELS...KNKVSVTCLVK DFLP..TDIT VEWQSNEHPEPEG.KYRTTEAQKDSD......GSYFLYSKLTVETDRW..QQGTTFTC VVMHEA.LHNHVM QKNVSHSP...GK
AJ302058,IGHG4 (G)QPREPQVYVLAPHRAELS...KNK?SVTCLVK DFYP..TDID IEWKSNGQPEPET.KYSTTPAQLDSD......GSYFLYSKLTVETNRW..QQGTTFTC AVMHEA.LHNHYT EKSVSKSP...GK
AJ312380,IGHG5 (G)QLRVPQVYVLAPHPDELA...KNTVSVTCLVK DFYP..PEID VEWQSNEHPEPEG.KYSTTPAQLNSD......GSYFLYSKLSVETSRW..KQGESFTC GVMHEA.VENHYT QKNVSHSP...GK
AJ312381,IGHG6 (G)ELQDPKVYILAPHREEVT...KNTVSVTCLVK DFYP..PDIN VEWQSNEEPEPEV.KYSTTPAQLDGD......GSYFLYSKLTVETDRW..EQGESFTC VVMHEA.IRHTYR QKSITNFP...GK
L49414 ,IGHM c (I)HQSPIQIFAIPPSFAGIFL..TKSAKLSCQVT NLGTY.DSLS ISWTRQNGEIL...KTHTNISESHPN......GTFSALGEATICVEDW..ESGDDYIC TVTHTD..LPFPL KQVISRPD
CH4
AJ305046,IGHE (G)KRLSPEVYVFLPPEEDQSS..KDKVTLTCLIQ NFFP..ADIS VQWLRNNVLIQTD.QQATTRPQKANGP....NPAFFVFSRLEVSRAEW..EQKNKFAC KVVHEA.LSQRTL QKEVSKDP...GK
L49414 ,IGHM c (A)VGKHPPSVYVLPPHREQLSL..RESASITCLVK GFLTP.PDVS VQWLQEGQPLSPD.KYVTSAPMLDPSP....QALYFVHSILTVSEEDW..SSGETYTC VVGHEA.LPHVVT ERTVDKSH...CKPTLYNVSLVMSDMASTCYCKPTLYNVSLVMSDMASTCY
Hinge
AJ302055,IGHG1 (E)PIPDNHQKVCDMSKCPKCP
AJ302056,IGHG2 (A)RVTPVCSLCRGRYPHPI
AJ312379,IGHG3 (E)PVLPKPTTPAPTVPLTTTVPVETTTPPCPCECPKCP
AJ302058,IGHG4 (V)IKECGGCPTCP
AJ312380,IGHG5 (V)VKGSPCPKCP
AJ312381,IGHG6 (V)IKEPCCCPKCP
c: cDNA sequence
Created: 10/06/2002
Last updated: 02/10/2025 13:10
Author: Delphine Guiraudou