Created: 12/11/2014
Last updated: 02/10/2025 13:10
Authors: Saida Saljoqi
The IGHC protein display numbering is according to the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN.
Letters in red correspond to amino acids which are polymorphic in the other alleles.
For C-REGIONs, letters in bold correspond to additional positions in the IMGT unique numbering.
For C-REGIONs, letters between parentheses correspond to amino acids resulting from the splicing.
N (Asn, asparagine) of potential N-glycosylation sites (NXS/T, where X is different from P), (N-linked glycosylation) is shown is green (site is underlined in CHS and in pages edited before 14/10/2009).
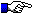 The hinge exon(s) are shown at the end of the protein display.
The hinge exon(s) are shown at the end of the protein display.
_____________________________________________________________________________________________________________________________________________________________________________________________
IGHC
genes
_____________________________________________________________________________________________________________________________________________________________________________________________
A AB B BC C CD D DE E EF F FG G CHS
--------------> ----------> ------> -------> -----------> -------> ---------->
1 10 15 16 20 23 30 36 39 41 45 77 84 85 89 96 97 104 110 118 121 130 140 150
7654321|........|....|123|...|...... ...|.AB....| |.....|1234567|......|12345677654321|..........|12|....... .....|....... ..|.........|.........|.........|
CH1
AY055778 ,IGHA1 c (V)DPKSPPVVFPLETCSSEN....QDPVVVACLVK GIFP....MPGV VYWDSDQASS....FTRTYNSIQSTGG......SFSFVSQLTMLSSQC..PLEKEFHC TAIYNN..LKATA NVT
NW_001794226 ,IGHA2 (A)TKTAPTIFPLNAYGSNS....EEQVSIGCLVR GYFP....EPVE VTWGNQASSA....VKTYPAVLESNG.......YYLVSSMLTLPANQC..PDDKTYQC NVKHY...DSSKT ANVKCR
NW_001794226 ,IGHD (V)GQRAPEVYPLYAGCGSPAPA.EDKVTLVCLAT NFSP....NDLA FTWNIKDDVK....NFPSVRRSDMT.........YIESSQLTFPASKW...DSETPTC TVKHN...QQKEK KIIVFKRQ
NW_001794226 ,IGHE (V)SSKAPSVFPLVPCCDGTD...SSAVTLGCLVT GYIP....EPVT VRWNSGDLVK....GVTTFPSVFDSQS.....GLYTMSSQVTVSQESW...QSQTFTC NVEQTATKTKINT EVYS
NW_001794226 ,IGHG1 (V)PPSIFPLTPSCKEEE...LSEVTVGCLVT GYSP....QPVH VKWSPPAFSS....SSKTFPAVLQSG......GLYTLSSQITLPTEKW...RSESVTC NVEHPA..SKTKI DKKV
NW_001705893 ,IGHG2 (A)AKVSPTVYPLIPSRKEKN...LSQVTLGCLVT GYFP....EPVS IKWSPETFSS....NSKTFPAILQPS......GLYTLSSQITVPEDNW...CGELFTC SVEHPA..TQKNT QKTV
NW_001794226 ,IGHM (A)SQQAPLLFPLVPSCGDSLNQ.ESSLAVGCLAK NFLP....NSIT LSWDFKNRTTVG..TEKFINYPTVLSG.....QAYSTTSQLMISATEVMMGQDDYVFC KAKHA....VGNK EIRVPLR
NW_001794226 ,IGHO (V)WKVAPSVFPLTPSCEEVE...LSEVTLGCLVT GYSP....DPVK VDWSPSFYNS....KAQTYPSILHPT......GLYSLSSKITVPAYNW...HNKVYTC KVTHTP..TNTII SGNV
CH2
AY055778 ,IGHA1 c CCKCSPKVLLHPPSLEGLFL..GKGANLTCVLK GLVDP...RETT FTWTRPNGDP....SQATTGNPVEEED.....GTYSLASVLEICAEEW..HQRDKFTC TVTHPK...MSPI TQTITKPP
NW_001794226 ,IGHA2 CCSNCPSVSVSLHPPSLESLFL..DKGANLTCELT GVSNV...KGVN FTWSPLSGTA....RPVDGPAVKDDK......GKYTITSTLEVCTDEW..MRGDKYTC TVSHPE..LPKPV TKTITKVS
NW_001794226 ,IGHD (D)QRMQPPRVHLLPPPCVGTSE..GGTVELICLLL QFKP....NSTE VQWLLNGRELNK..PAPTLFAADQDG.......SFMARSLLRISSSNW..EGGDTFTC RVTHPA..LKEGP ELYNSSKCS
NW_001794226 ,IGHE (D)CSKDPIPPTVKLLHSSCDPRGDS.QASIELLCLIT SYSP....AGIQ VDWLVDGQKAEN..LFPYTAPPKREGN.....RSFSSHSEVNITQDQW..LSGKTFTC QVTHLA..DKKTY QDSARKCA
NW_001794226 ,IGHG1 (S)SAAVFIFPPKPKDFLSV.AGTPKVTCVVV DLGFEDKDESPV VTWYQGDKELP...KTRMLEPPPKEQR....NGTYRFVSERDVSSKDW..LDQKVFKC KVQHKN..FPSPI TKSISHTA
NW_001705893 ,IGHG2 (V)LCGPAVFLVPPRPKDLLSE.GGKPKIICVAT GLRDE..EKDAR VKWYKNGTLFP...NSQNAQTSSDTIW.....NGTRVSSKLSVTPEDW..KSDAEFRC EVEHKL..FPTSL KKAISHDK
NW_001794226 ,IGHM (V)AQPSRPNVSLHIPTREEFLGP.ELEGTIICQAK HFNP....KEIS LKWLKNGELLNG..GFTTEGPTLDGS......KTYQLSSSLTVSDTDW..FKATDFTC VVEHP....KTHF IEMRNASYKCAG
NW_001794226 ,IGHO (1) VTPAMKSPPAVRVVHSSCLPNGDA.EATIQLLCLIS DFSP....AKIE VKWLEDDEEQDG..FYVSESKRENNG.......KFSAYSEFNITQGEW..LTGKTFTC SVRHMA..SKKDI QDYARSCK
CH3
AY055778 ,IGHA1 c (G)PLNRPEVHLLAPSTEELAL..NEMATLTCLVR GFNP....PDLL VKWLKGGQEVSQT.DYVTSSPQREASEG..SASTFFLYSTLRVPTSEW..KEGENYSC VVGHEA.LPLNFT QKTIDHST...GKPSTVNVSLVMSDTAGTCY
NW_001794226 ,IGHA2 (G)PLFRPEVHLLAPTAEELAL..NELATLTCLVR GFSP....KELM VKWLKGGQEVPRK.DYVTGSPQREVSEG...SATFFLYSTLRVQTSTW..KEGENFSC VVGHES.LPLNFT QKTFDHST...GKPSTVNLTVVMSDAANTCY
NW_001794226 ,IGHD (A)CHSHATAMTLYLFAPSYDELVRG.NGKGHATCLVL GYDL....ASVQ ISWQVNGVVTEGQ.TGVTRHESNGTES.....RTSTQTISLADWKA......GRRYIC RAKHPCVKEVTQE LSTFDP
NW_001794226 ,IGHE (D)SDPRGITVFLTPPSPTDLYI..SKTPKLTCLII DLVST...EGME VTWSRESGTPL...SAESFEEQKQFN......GTMSFISTVPVNIQDW..NEGESYTC RVAHPD..LPSPI IKTVTKLP
NW_001794226 ,IGHG1 (G)TRKAPEAYVFPPHRDEMN...KNSVSITCLVL DFYP....DAIA IDWQHNDRPVP...EGDYATTLPQKN.....GNSYFLYSKLTVQKTEW..NQGGSFTC SVRHEA.LPQKFL QKTVSKTS...GK
NW_001705893 ,IGHG2 (N)TERKTPEAYIFSPHTEELS...KESVSITCLVK GFLP....QDIS IAWQHKGKEMK...EEEYSTTPPQYR.....NGSYFLYSKLTVTKKDW..ETGESFSC LVYHDS..RSDTL KKTVSKNT...GN
NW_001794226 ,IGHM (V)PSSSIKISTTPPSFAGIFT..TKSAKLVCSVT GMSTS...DSLS IVWTQYEGKEL...TTKTSVSKIQPD......GTFSATGEASVCAEDW..QSGKRFTC TVSHAD..LPAPQ SAYISKQK
NW_001794226 ,IGHO (D)INLIQNLKVSVSPPNPTDLFI..ARTPSLTCLVA SLPDS...EGIK LQWIHKGIER....PASPIKIMKQPD......GTFSAESTLSITLPEW..MEGESFTC KVQHPD.FPSIVE KAISKPL
CH4
NW_001794226 ,IGHD (K)TPTKKPSVSVSRVFSEDSPGE..EDHPTLLCNIR GFYP....KELS VSWENNSHPLP...RTLYSSGPVVAT.....GNEAFSTYSLLRVGPGE...RGGAYTC VVMHPS.TDRPTR SNKVSF
NW_001794226 ,IGHE (G)KRLAPEVYAFPPHQAEVSH..GDSLSLTCLIR GFYP....ENIS VRWLLDNKPLPTE.HYRTTKPLKDQGP....DPAYFLYSRLAVNKSTW..EQGNVYTC QVVHEALPSRNTE RKFQHTS....GN
NW_001794226 ,IGHM (E)VNLHSPAVYVLPPASDQLSL..RESATLSCLVK SFSP....ADVM VQWLHKGQPVPQD.KYTVSAPMPEPQS....PNLYFAYSTLTVAEEEW..SAGDSFTC VVVHEA.LPLYVM ERTVDKST...GKPTHINVSLVMSDTANTCY
NW_001794226 ,IGHO (G)KSLAPAVYVFPPHMDELAQ..KDTLSLTCLAK SFFP....ADIA VQWLHNDEDVEED.HFSVTKPQKDLTG....QGTFFLYSKLDIQKSNW..KRGDSFTC MVGHEALKRYSI. QKTVRQNP...GK
CH5
NW_001794226 ,IGHD (D)WALSRAPEVSITGHPACNAPG..AETSVFRCSAS NFFP....AGAE ILWLVKGQEQPG..LTQTITRSRDGR........YSASLELSVPNAQW..NQLAEFTC RVSHAG..ATQQK NLSKCA
CH6
NW_001794226 ,IGHD (V)CQDSTPQPTLYLLPPPLEGLVL..RAEALLTCLVV GYQL....DQAT LTWEVDKKPLAG..SPSVRALVTHDN......GTQSLRSHLNLSRTDW..EAAHTVLC RVSHRC..LFAPQ EETLQPLR
CH7
NW_001794226 ,IGHD (G)ADPPRKLSIHVLHPSLPQSSA..EASAWLLCEVS NFSP....AEIF LTWWENETSVSPS.WFTTTAPFAQPGN.....AMYSTQSLLRIPASQG..HMASTYTC MVGHVSSPSMLNI SRNGVF
CH8
NW_001794226 ,IGHD (D)FLEPARPLVTAFHEPSG..SSREKLVCLAT DFRP....KDID MKWEVRGQERRC..SDSLPSKALGNG.......MFQQSCALLLSLEEW..TQPETYTC VVNHSS..DTAAV RRELTST
CH9
NW_001794226 ,IGHD (Q)CPAWSPTAHTLPPSFEWLLQ..NETVVLTCVTD MF.......NAS VEWTAGEELAAL..ETVENKKEKFPN......GSSWLFSQLKVNFSVW..NSTESFSC QVKDGQ..SELRV NVTHIP
CH10
NW_001794226 ,IGHD (G)PVKAPKVYLFPPPTEEMSR..GPSLTLVCLIK DFYP....ASIL VQWQEGSAVLETQAAQSQNPSCDHQS......RRCSAVSKLEIPKSRW..MARTRYHC QVFHIS..RRGIL IRNASTHY (D)HPAEDSGS
M
NW_001794226 ,IGHA2 (G)PECYCPTTVACCSFPKLPVVDEGGVEETDQESTLWPTSVTFLTLFLLSLFYSTTVTLFKVKPLSPIKY
M1 M2
NW_001794226 ,IGHD (D)PRVIDFTFKDENNDDYTEMDESNNVWPTVSTFVALFLLTLLYSGFVTFIK VK
NW_001794226 ,IGHE (D)LIREDDCSEAENEELDGLWTTIYVFITLFLLSVGYSATVTLFK VKWIFATVLQLKRSNVHDYKNMIQQGA
NW_001794226 ,IGHG1 (E)LILDENCAEAGDEELEGLWTTISVFITLFFLSVCYSATVTLFK VKWIFSTVVDLKPQMLPDYRNMIDQGA
NW_001705893 ,IGHG2 (G)LILDENCTEADSEELEGLWTTISVFITLFFLSVCYSATITLFK VKWIYSSVVQLKPQMLPDYRNMINHEV
NW_001794226 ,IGHM (E)GEVNAEEEGFENLWTTASTFIVLFLLSLFYSTTVTLFK VK
NW_001794226 ,IGHO (G)LIPGENCTEAGDGELDGLWTTISIFITLFLLTVCYSATITLVK VRWLFSSMMQIKQTRSIHNYRNMILPGA
CONNECTING-REGION | TRANSMEMBRANE-REGION | CYTOPLASMIC-REGION
Hinge
AY055778 ,IGHA1 (C)PPVPQ
NW_001794226 ,IGHA2 (S)PVSNCGCE
NW_001794226 ,IGHG1 (E)PGPSNPPHCP
NW_001705893 ,IGHG2 (G)PRFPGSNSCP
NW_001794226 ,IGHO (V)APAPPTPQTTTPV
c: cDNA sequence
| (1) | The determination of CH2 is arbitrarily based on the 5' delimitation of IGHE CH2. |