- Homo sapiens: RAET1E
IMGT RPI entry from gene to protein for Homo sapiens RAET1I
IMGT gene name and definition
- IMGT gene name: Homo sapiens RAET1I
- IMGT gene definition: retinoic acid early transcript 1 I, UL16 binding protein 1 (ULBP1)
Chromosomal localization
- Chromosome: 6
- Chromosomal localization: 6q25.1
Locus representation
Homo sapiens RAET1I locus 6q25.1
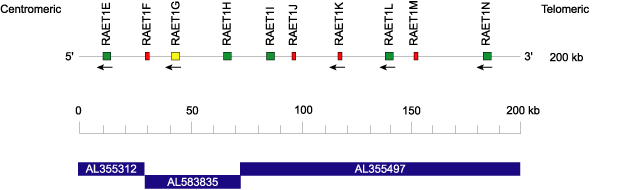
The human RAET1 locus [1] located at 6q25.1 spans 200 kilobases from 150,230 to 150,430 kb (Ensembl, 09/06/2004).
Clones (in blue) are in forward (FWD) orientation on the chromosome. They overlap by about 100 base pairs of complete similarity. An additional 100 base pairs are dissimilar probably for cloning reasons.
Gene exon/intron organization
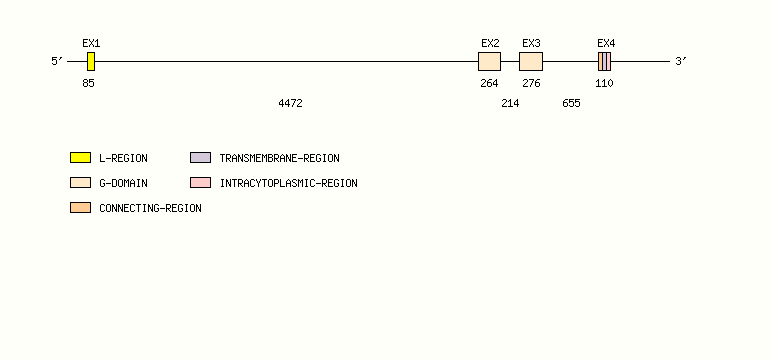
Legend:
- Length of the exons and introns are in base pairs.
- Colors are according to IMGT Color menu for regions and domains
Number of alleles: 2
IMGT reference alleles
| Allele name | Gene functionality | IMGT reference sequences | ||
|---|---|---|---|---|
| Clone names | Accession numbers | Molecule type | ||
| RAET1I*01 | F | RP11-472G23 | AL355497 [1] | gDNA |
| RAET1I*02 | F | AF304377 | cDNA | |
IMGT reference sequences (in FASTA format) for the allele(s): RAET1I*01 to RAET1I*02
- Nucleotide sequence in FASTA format with gaps according to the IMGT unique numbering
- Amino acid sequence in FASTA format with gaps according to the IMGT unique numbering
- Nucleotide sequence in FASTA format without gaps
- Amino acid sequence in FASTA format without gaps
Other sequences from the literature
| Allele name | Gene functionality | IMGT reference sequences | ||
|---|---|---|---|---|
| Clone names | Accession numbers | Molecule type | ||
| RAET1I*01 | F | AF346416 | cDNA | |
| AF425265 | cDNA | |||
| RAET1I*02 | F | AB052907 | cDNA | |
Corresponding protein database accession numbers
| Allele name | Accession numbers | Number of amino acids | Protein isoform | |
|---|---|---|---|---|
| Nucleotide databases | Protein databases | |||
| RAET1I*01 | AF346416 | Sw: Q9BZM6 | 244 aa | |
| AF425265 | 239 aa | (1) | ||
| RAET1I*02 | AF304377 | Tr: Q9BZM6 | 244 aa | |
| AB052907 | 244 aa | |||
| NM_025218 | NP_079494 | 244 aa | ||
- (1) in 5', 13 nucleotides or 5 amino acids missing.
- [1] Radosavljevic, M. et al., Genomics, 79, 114-123 (2002). PMID:11827464
Genomic sequence
Coding region sequence
The coding region (CODING-REGION)
sequence starts from INIT-CODON
(or its encoded amino acid) to STOP-CODON
(not included).
The nucleotide sequence is extracted from cDNA (c) sequences and/or is
built by artificial exon joining from genomic DNA (g) sequences.
The amino acid sequence is the translation of the nucleotide sequence.
RAET1I*01: AL355497(g) AF346416(c)
Nucleotide sequence
1 atggcagcgg ccgccagccc cgcgttcctt ctgtgcctcc cgcttctgca cctgctgtct 61 ggctggtccc gggcaggatg ggtcgacaca cactgtcttt gctatgactt catcatcact 121 cctaagtcca gacctgaacc acagtggtgt gaagttcaag gcctggtgga tgaaaggcct 181 tttcttcact atgactgtgt taaccacaag gccaaagcct ttgcttctct ggggaagaaa 241 gtcaatgtca caaaaacctg ggaagaacaa actgaaacac taagagacgt ggtggatttc 301 cttaaagggc aactgcttga cattcaagtg gagaatttaa tacccattga gcccctcacc 361 ctgcaggcca ggatgtcttg tgagcatgaa gcccatggac acggcagagg atcttggcag 421 ttcctcttca atggacagaa gttcctcctc tttgactcaa acaacagaaa gtggacagca 481 cttcatcctg gagccaagaa gatgacagag aagtgggaga agaacaggga tgtgaccatg 541 ttcttccaga agatttcact gggggattgt aagatgtggc ttgaagaatt tttgatgtac 601 tgggaacaaa tgctggatcc aacaaaacca ccctctctgg ccccaggcac aacccaaccc 661 aaggccatgg ccaccaccct cagtccctgg agccttctca tcatcttcct ctgcttcatt 721 ctagctggca gatga
Amino acid sequence
1 MAAAASPAFL LCLPLLHLLS GWSRAGWVDT HCLCYDFIIT PKSRPEPQWC EVQGLVDERP 61 FLHYDCVNHK AKAFASLGKK VNVTKTWEEQ TETLRDVVDF LKGQLLDIQV ENLIPIEPLT 121 LQARMSCEHE AHGHGRGSWQ FLFNGQKFLL FDSNNRKWTA LHPGAKKMTE KWEKNRDVTM 181 FFQKISLGDC KMWLEEFLMY WEQMLDPTKP PSLAPGTTQP KAMATTLSPW SLLIIFLCFI 241 LAGR*
IMGT notes:
S 212 (underlined) is a potential GPI anchor (potential cleavage site ω, ω+1, ω+2 at 212-214).
Legend:
- Colors are according to IMGT Color menu for regions and domains
- Nucleotide and amino acid letters (in bold) in purple, green or blue are according to IMGT Color menu for splicing types.
IMGT links
IMGT Repertoire
- Chromosomal localization:
- Alignment of alleles:
- Protein displays:
- IMGT Collier de perles: G-LIKE-DOMAINs
External links
Nomenclature
- HGNC: 14893
Genome databases
- Entrez Gene: 80329
- GENATLAS:
- GeneCards: GC06P150316
- GDB: 11502788
- OMIM: 605697



