- Citing IMGT/StatClonotype:
- Aouinti S., Giudicelli V., Duroux P., Malouche D., Kossida S., Lefranc M-P.
Front. Immunol. 7:339 (2016).
doi:10.3389/fimmu.2016.00339
PMID: 27667992
Abstract
Full

- Aouinti, S., Malouche, D., Giudicelli, V., Kossida, S., Lefranc, M-P.
PLoS One, 10(11):e0142353 (2015).
doi:10.1371/journal.pone.0142353
PMID: 26540440
Abstract
Full

- IMGT/StatClonotype version:
- 1.0.4 (January 25th 2021)
-
Introduction
- IMGT/StatClonotype [1], developed by LIGM
(Montpellier University, CNRS) and part of IMGT®, the international ImMunoGeneTics information system® (http://www.imgt.org) [2] is a standalone IMGT® tool for statistical analysis of sets from IMGT/HighV-QUEST output [3,4], on the Web since June 2016.
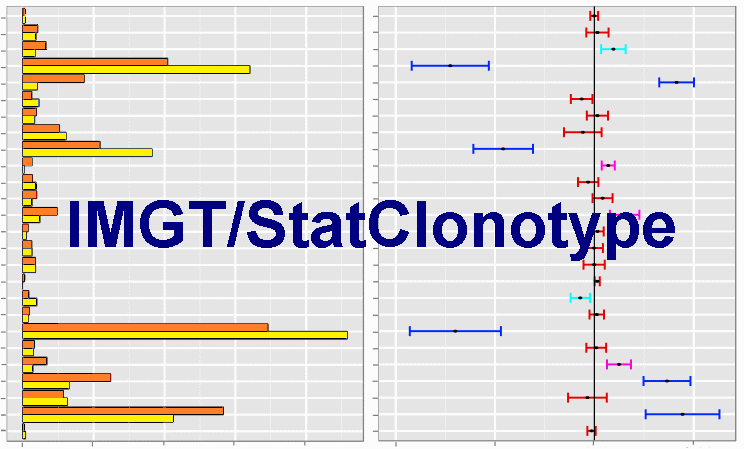
 IMGT/StatClonotype works only with uploaded files (.txt format) from the IMGT/HighV-QUEST statistical analysis output ('stats_xxx' file available in 'data' directory of the IMGT/HighV-QUEST statistical analysis output where 'xxx' is the batch name and the locus type).
IMGT/StatClonotype works only with uploaded files (.txt format) from the IMGT/HighV-QUEST statistical analysis output ('stats_xxx' file available in 'data' directory of the IMGT/HighV-QUEST statistical analysis output where 'xxx' is the batch name and the locus type).
To get these files, you therefore need to launch the IMGT/HighV-QUEST statistical analysis
(IMGT/HighV-QUEST Documentation).- IMGT/StatClonotype uses a generic statistical procedure [1] for identifying significant changes in IG and TR differences of proportions of IMGT clonotypes (AA) diversity and expression [5].
It uses the IMGT gene and allele nomenclature [6,7,8] based on IMGT-ONTOLOGY [9] and IMGT standards in immunoinformatics [10].
- IMGT/StatClonotype is a tool incorporated in a new R package ('IMGTStatClonotype') to perform pairwise comparison of sets from IMGT/HighV-QUEST output
through a user-friendly web interface implemented using Shiny framework [11] in users' own browser.
- IMGT/StatClonotype source is distributed under the GNU Lesser General Public License.
IMGT/StatClonotype installation
IMGT/StatClonotype should be installed following the IMGT/StatClonotype Documentation
Package for users acquainted with R software
Example files for testing IMGT/StatClonotype
| Examples | | Files | Locus | Description | Last modified |
|---|
|
|---|
| Example 1 | ![[TXT]](/StatClonotype/images/text.gif) | file 1 MID1.txt | TRB | CD4- T cells | 16-Dec-2015 |
![[TXT]](/StatClonotype/images/text.gif) | file 2 MID2.txt | TRB | CD4+ T cells | 16-Dec-2015 |
| Example 2 | ![[TXT]](/StatClonotype/images/text.gif) | file 1 S1.txt | IGH | Memory IgD+ | 16-Dec-2015 |
![[TXT]](/StatClonotype/images/text.gif) | file 2 S2.txt | IGH | Memory IgD- | 16-Dec-2015 |
MID1.txt, MID2.txt (example 1) and S1.txt, S2.txt (example 2) are from IMGT/HighV-QUEST statistical output, available in the NCBI Sequence Read Archive under the accession code SRX326382 and SRP037774, respectively.
References
[1]
Aouinti, S., Malouche, D., Giudicelli, V., Kossida, S., Lefranc, M-P.
IMGT/HighV-QUEST statistical significance of IMGT clonotype (AA) diversity per gene for standardized comparisons of next generation sequencing immunoprofiles of immunoglobulins and T cell receptors.
PLoS One 10(11):e0142353. (2015)
Free Article
PMID: 26540440.
[2]
Lefranc, M-P., Giudicelli, V., Duroux, P., Jabado-Michaloud, J., Folch, G., Aouinti, S. et al.
IMGT®, the international ImMunoGeneTics information system® 25 years on.
Nucleic Acids Res. 43(Database issue):D413-22. (2015)
Free Article
PMID: 25378316.
[3]
Alamyar, E., Giudicelli, V., Shuo, L., Duroux, P. and Lefranc, M.-P.
IMGT/HighV-QUEST: the IMGT® web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing.
Immunome Res. 8:1:2. (2012)
PMID: 22647994
 .
.
[4]
Alamyar, E., Duroux, P., Lefranc, M.-P. and Giudicelli, V.
IMGT tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT HighV-QUEST for NGS.
Methods Mol. Biol. 882:569-604. (2012)
PMID: 22665256.
[5]
Li, S., Lefranc, M.-P., Miles, J.J., Alamyar, E., Giudicelli, V., Duroux, P., et al. IMGT/HighV-QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling.
Nature Comm. 4:2333. (2013)
Open access
PMID: 23995877.
[6]
Lefranc, M-P. and Lefranc, G.
The T cell receptor FactsBook.
Academic Press, London, UK (398 pages), 2001.
[7]
Lefranc, M-P. and Lefranc, G.
The Immunoglobulin FactsBook.
Academic Press, London, UK (458 pages), 2001.
[8]
Giudicelli, V., Chaume, D., Lefranc, M-P.
IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes.
Nucleic Acids Res. 33(Database issue):D256-61. (2005).
[9]
Giudicelli, V., and Lefranc M-P.
IMGT-ONTOLOGY 2012.
Frontiers in Bioinformatics and Computational Biology.
Online access
Front. Genet. 3:79. (2012).
PMID: 22654892
[10]
Lefranc, M-P.
Immunoglobulin (IG) and T cell receptor genes (TR): IMGT® and the birth and rise of immunoinformatics.
Front Immunol. 5:22. (2014).
doi: 10.3389/fimmu.2014.00022.
Open access
PMID: 24600447
[11]
Beeley, C. et al. (2013)
Web application development with R using Shiny.
Packt Publishing Ltd, 110 pages.
- Created:
- Wednesday, 12 August 2015
- Last updated:
- Monday, 25 January 2021
- Author:
- Safa Aouinti safa.aouinti@yahoo.fr
- Contacts:
- Safa Aouinti safa.aouinti@yahoo.fr, Sofia Kossida Sofia.Kossida@igh.cnrs.fr and Marie-Paule Lefranc Marie-Paule.Lefranc@igh.cnrs.fr
 WELCOME !
WELCOME ! WELCOME !
WELCOME !
 IMGT/StatClonotype works only with uploaded files (.txt format) from the IMGT/HighV-QUEST statistical analysis output ('stats_xxx' file available in 'data' directory of the IMGT/HighV-QUEST statistical analysis output where 'xxx' is the batch name and the locus type).
IMGT/StatClonotype works only with uploaded files (.txt format) from the IMGT/HighV-QUEST statistical analysis output ('stats_xxx' file available in 'data' directory of the IMGT/HighV-QUEST statistical analysis output where 'xxx' is the batch name and the locus type).![[ ]](/StatClonotype/images/compressed.gif)
![[TXT]](/StatClonotype/images/layout.gif)
![[TXT]](/StatClonotype/images/text.gif)
![[TXT]](/StatClonotype/images/text.gif)
![[TXT]](/StatClonotype/images/text.gif)
![[TXT]](/StatClonotype/images/text.gif)