IMGT/PRIMER-DB contains information on primers and combinations of primers described as "sets" and "couples".
 The definition, taxonomy species, classification (IMGT group,
IMGT subgroup, IMGT gene,
IMGT allele) and label description of an IMGT/PRIMER-DB primer
are those of the IMGT/LIGM-DB reference sequence, and not those of the PCR
amplification products.
The definition, taxonomy species, classification (IMGT group,
IMGT subgroup, IMGT gene,
IMGT allele) and label description of an IMGT/PRIMER-DB primer
are those of the IMGT/LIGM-DB reference sequence, and not those of the PCR
amplification products.
This provides the following advantages for the data standardization:
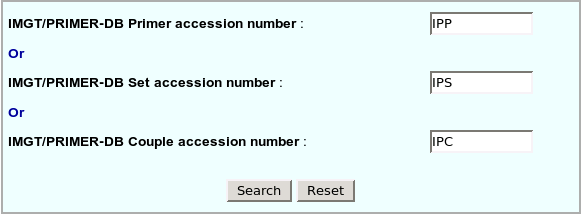
| IMGT/PRIMER-DB Primer accession number | Unique identifier of each primer in IMGT/PRIMER-DB.
Consists of 3 letters "IPP" (for IMGT/PRIMER-DB Primer) and 6 numbers. |
| IMGT/PRIMER-DB Set accession number | Unique identifier of each set in IMGT/PRIMER-DB.
Consists of 3 letters "IPS" (for IMGT/PRIMER-DB Set) and 6 numbers. |
| IMGT/PRIMER-DB Couple accession number | Unique identifier of each couple in IMGT/PRIMER-DB.
Consists of 3 letters "IPC" (for IMGT/PRIMER-DB Couple) and 6 numbers. |
Each IMGT/PRIMER-DB primer, set or couple, identified by an IMGT/PRIMER-DB accession number is described in an IMGT/PRIMER-DB Primer cards, IMGT/PRIMER-DB Set cards and IMGT/PRIMER-DB Couple cards, respectively.
 Search can be made directly by the IMGT/PRIMER-DB Primer
accession number, IMGT/PRIMER-DB Set accession number or IMGT/PRIMER-DB Couple
accession number. Search can be made directly by the IMGT/PRIMER-DB Primer
accession number, IMGT/PRIMER-DB Set accession number or IMGT/PRIMER-DB Couple
accession number. |
 |
| This query gives access to the corresponding IMGT/PRIMER-DB Primer card, IMGT/PRIMER-DB Set card or IMGT/PRIMER-DB Couple card. |
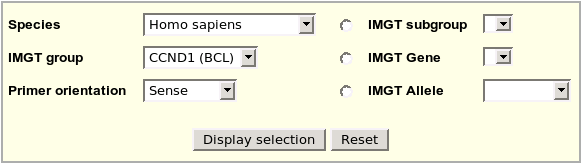
 Primers may be selected on the following criteria: Species,
IMGT group and Primer orientation Primers may be selected on the following criteria: Species,
IMGT group and Primer orientation |
 |
|
|
The resulting page displays:
A button allows to classify that list by IMGT/PRIMER-DB Primer accession numbers, IMGT subgroups, IMGT gene names or IMGT allele names.
| IMGT/PRIMER-DB Primer accession number: | 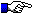 Click on t>e IMGT/PRIMER-DB Primer accession number to get access to
the IMGT/PRIMER-DB Primer card Click on t>e IMGT/PRIMER-DB Primer accession number to get access to
the IMGT/PRIMER-DB Primer card
|
| Species: | Latin name is according to the IMGT Taxonomy (IMGT Repertoire). |
| IMGT/PRIMER-DB Primer orientation: | "Sense" or "Antisense" is by comparison with the gene transcription orientation. |
| IMGT group | |
| IMGT subgroup: | Only for variable genes |
| IMGT gene name: | Gene name is according to the IMGT nomenclature: Nomenclature (IMGT Index), IMGT gene nomenclature (IMGT Scientific chart), and as used in IMGT Repertoire (Germline gene tables) and IMGT/GENE-DB. |
| IMGT allele name: | Allele name is according to the IMGT nomenclature: Allele (IMGT Index), IMGT allele nomenclature (IMGT Scientific chart), and as used in IMGT Repertoire (Alignments of alleles) and IMGT/GENE-DB. |
| - IMGT/PRIMER-DB Primer accession number: Unique identifier of each primer in IMGT/PRIMER-DB | |
| - IMGT/PRIMER-DB entry date | |
| - IMGT/PRIMER-DB update | |
| - IMGT/PRIMER-DB Primer definition: Automatically composed of the species name followed by the IMGT gene and allele name and the primer orientation. | |
| - Bibliographic references: 3 possibilities: | |
|
|
| - Catalogue comments | |
| - IMGT/PRIMER-DB Primer sequence: Given in bold CAPITAL letters. | |||||||
|
|||||||
| - IMGT/PRIMER-DB Sequence length: In number of nucleotides. | |||||||
| - IMGT/PRIMER-DB Primer orientation: "Sense" or "Antisense" (by comparison with the gene transcription orientation). | |||||||
| - IMGT/LIGM-DB reference sequence Accession number: | |||||||
Unique IMGT/LIGM-DB
accession number of the IMGT reference sequence which allowed the definition (in PRIMER CATALOGUE),
classification (in PRIMER CLASSIFICATION) and the label description (in PRIMER DESCRIPTION) of the primer. Click on the IMGT/LIGM-DB reference sequence Accession number to access the IMGT/LIGM-DB flat file. Click on the IMGT/LIGM-DB reference sequence Accession number to access the IMGT/LIGM-DB flat file.
|
|||||||
| - IMGT/LIGM-DB reference sequence Primer position: Primer position (start-end) in the IMGT/LIGM-DB flat file reference sequence. | |||||||
| - Characteristics comments | |||||||

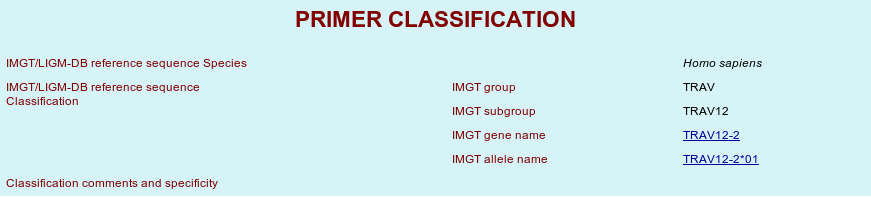
| - IMGT/LIGM-DB reference sequence Species: Latin name of species related to the primer, sometimes followed by common name between brackets. | |
| - IMGT/LIGM-DB reference sequence Classification: CLASSIFICATION comprises IMGT group, IMGT subgroup or cluster, IMGT gene name and IMGT allele name. | |
 Click on the IMGT gene name to view IMGT Colliers de Perles. Click on the IMGT gene name to view IMGT Colliers de Perles.
 Click on the IMGT allele name to view IMGT Alignments of alleles. Click on the IMGT allele name to view IMGT Alignments of alleles.
|
|
| - Classification comments and specificity: Specificity as described experimentally or deduced from sequence comparison, with a reference for the source of information. | |
| - IMGT/PRIMER-DB Primer labels: IMGT standardized labels used to describe the primer. | |
| For V-DOMAIN AND C-DOMAIN primers, amino acids are described with the IMGT unique numbering for V-DOMAIN and IMGT unique numbering for C-DOMAIN, respectively. Example: "AA_IMGT 1..8". | |
| - Differences between the IMGT/LIGM-DB reference sequence and the primer sequence. | |
| - Restriction enzyme name: Restriction site sequence and enzyme name: Example: "GAATTC EcoRI". | |
| - Restriction site position: Restriction site position in the primer. | |
| - Purification methods | |
| - Keywords | |
| - Description comments | |

|
|
Provides the list of the accession numbers of the IMGT/PRIMER-DB sets to which a
primer belongs. Click on the IMGT/PRIMER-DB Set accession number to
access the corresponding IMGT/PRIMER-DB Set card. Click on the IMGT/PRIMER-DB Set accession number to
access the corresponding IMGT/PRIMER-DB Set card.

|
|
| - IMGT/PRIMER-DB Set accession number: Unique identifier of each set in IMGT/PRIMER-DB |

| - IMGT/PRIMER-DB Set orientation: "Sense" or "Antisense". Defined by the primer orientation, common to all primers in the set. |

| - Species: Species Latin name: Defined by the primer species, common to all primers in the set. | |
| - IMGT group: Defined by the primer IMGT group, common to all primers in the set. |

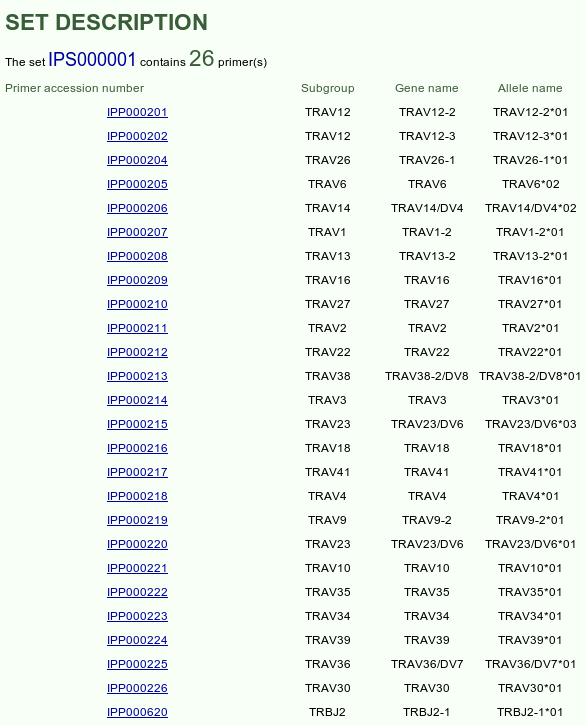
Provides the list of the IMGT/PRIMER-DB Primer accession numbers of all primers
contained in the selected IMGT/PRIMER-DB set, with IMGT subgroup, IMGT gene name and
IMGT allele name information. Click on an IMGT/PRIMER-DB Primer accession number to access
the corresponding IMGT/PRIMER-DB Primer card. Click on an IMGT/PRIMER-DB Primer accession number to access
the corresponding IMGT/PRIMER-DB Primer card. |
Provides the list of the accession numbers of the IMGT/PRIMER-DB couples to which a
set belongs.
 Click on the IMGT/PRIMER-DB Couple accession number to
access the corresponding IMGT/PRIMER-DB Couple card. Click on the IMGT/PRIMER-DB Couple accession number to
access the corresponding IMGT/PRIMER-DB Couple card.
|
 Click on an IMGT/LIGM-DB Accession number to access the flat file in IMGT/LIGM-DB.
Click on an IMGT/LIGM-DB Accession number to access the flat file in IMGT/LIGM-DB.

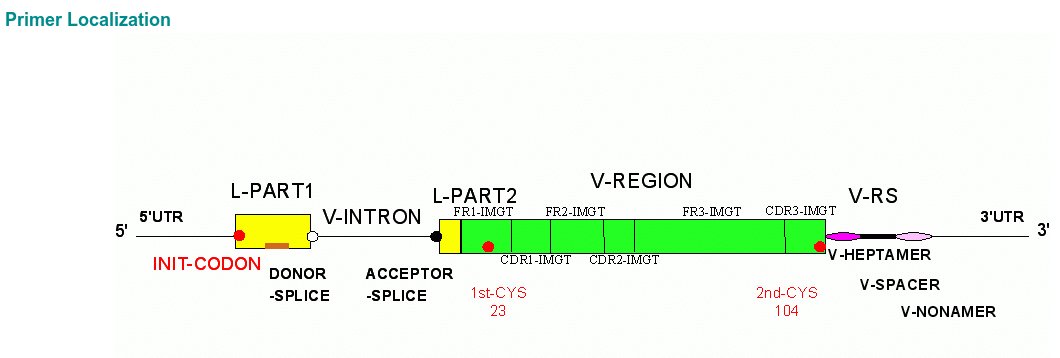
The localization of the oligonucleotides will be done on the gene sequences characterized in IMGT/GENE-DB (IMGT/GENE-DB is the IMGT genome database of immunoglobulins and receptors T).
The localization take into account information (annotations, "labels", positions according to single classification IMGT) which will be extracted from IMGT/LIGM-DB (IMGT/LIGM-DB is the IMGT sequence database of the immunoglobulines and the receptors T).
 Click on the IMGT/PRIMER-DB "Primer Localization" button to get
access to the IMGT/PRIMER-DB Primer Localization page.
Click on the IMGT/PRIMER-DB "Primer Localization" button to get
access to the IMGT/PRIMER-DB Primer Localization page.
The IMGT/PRIMER-DB Localization provides:
 Click on the IMGT/LIGM-DB Accession number to
access the flat file in IMGT/LIGM-DB.
Click on the IMGT/LIGM-DB Accession number to
access the flat file in IMGT/LIGM-DB.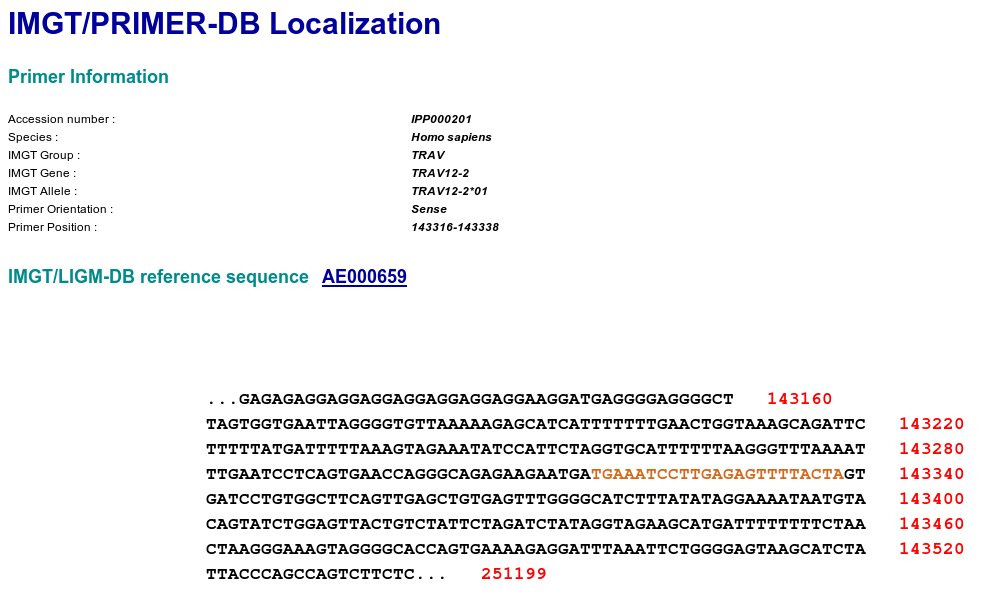
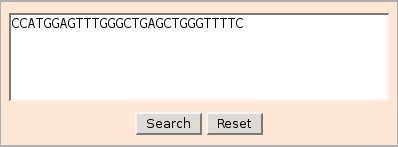
The comparison sequence with all the IMGT/Primer-DB sequences is made using fasta35*, a program which compares a protein sequence
to a protein sequence database or a DNA sequence to a DNA sequence database using the FASTA algorithm (Pearson and Lipman, 1988, Pearson, 1996).
Search speed and selectivity are controlled with the ktup(wordsize) parameter. For protein comparisons, ktup = 2 by default;
ktup =1 is more sensitive but slower. For DNA comparisons,
ktup=6 by default; ktup=3 or ktup=4 provides higher sensi-
tivity; ktup=1 should be used for oligonucleotides (DNA
query lengths < 20).
- The IMGT/PRIMER-DB Primer accession number
- The Species
- The Smith-Waterman Score
- The Percentage identity
- The Overlap
- The alignment

-ktup: Search speed and selectivity are controlled with the ktup(wordsize) parameter. ktup=1 should be used for oligonucleotides (DNA query lengths < 20).
-c: Set to 1 to optimize every sequence in a database.
-A: Force Smith-Waterman alignments for fasta3 DNA sequences. By default, only fasta3 protein sequence comparisons use
Smith-Waterman alignments.
-b: Number of sequence scores to be shown on output. Set to 10.
-E: Limit the number of scores and alignments shown based on the expected number of scores. Used to override the expectation
value of 10.0 used by default.Set to 10.
-m: Specify alignment type: 0, 1, 2, 3, 4, 5, 6, 9, 10. Set to 0.
-z: Set to 0. z 0 estimates the significance of the match from the mean and standard deviation of the library scores, without correcting for
library sequence length.