Multigene immunoglobulin heavy constant (IGHC) deletions have been found in healthy individuals,
either homozygous for the same deletion or heterozygous for two different deletions, in the IGH locus.
They represent gene copy number variations (CNV) and correspond to different haplotypes.
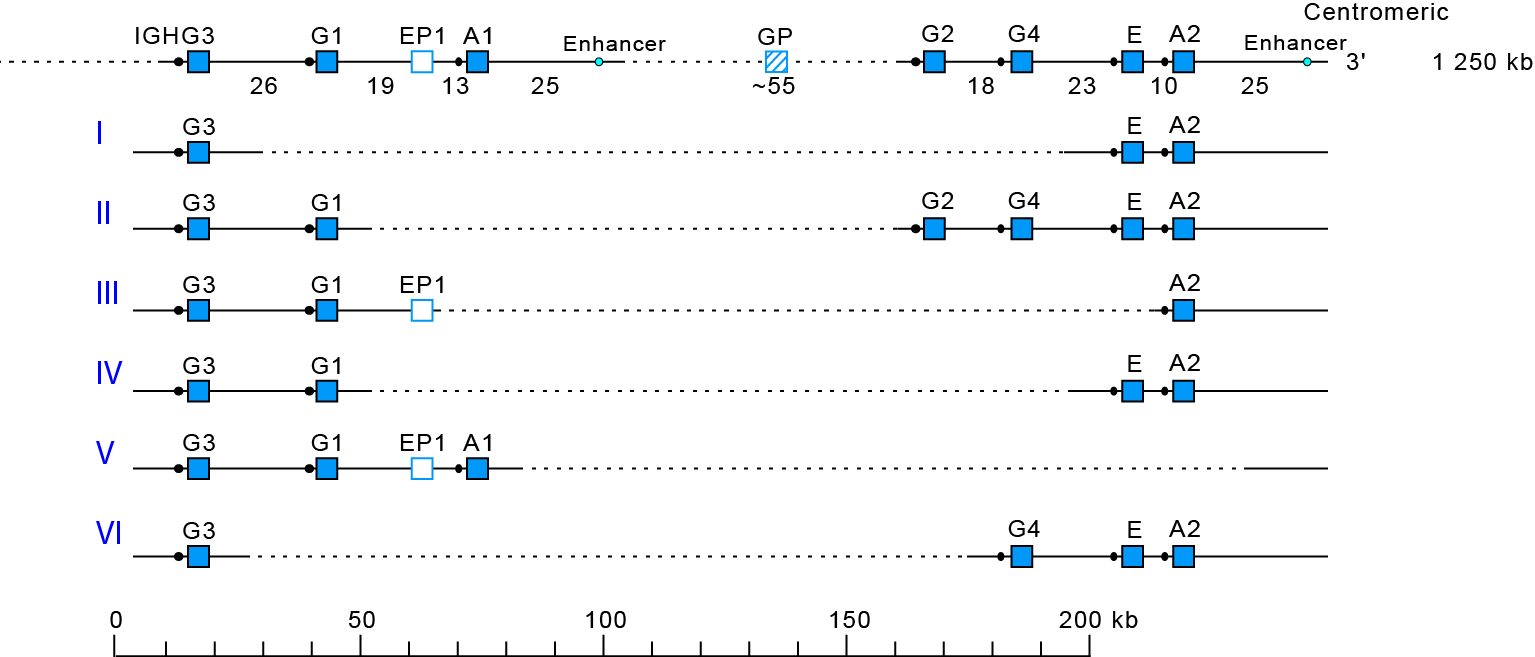
Deletions I [1,2] and II [3] contributed to order the Homo sapiens IGHC gene in the IGH locus.
These multigene IGHC deletions involve highly homologous spots of recombination [4], as also described for deletion III [5].
Six different multigene haplotypes have been identified, designed I to VI according to the chronogical order in which they were found :
| Origine | Number of cases | Proband | Absent immunoglobulins | Genotypes with reference to the type of deletion | References |
|---|---|---|---|---|---|
| Family HASS Tunisia |
3 | TAK3 (1) | IgG1, IgA1, IgG2, IgG4 | I/I | Lefranc M-P. et al. (1982) [1] Lefranc G. et al. (1983) [2] |
| Family TOU Tunisia |
3 | TOU II-I TOU II-4 (EZZ) TOU II-5 |
IgG1, IgA1, IgG2, IgG4 | I/I | Lefranc M-P. et al. (1982 [1], 1983 [3]) |
| 2 | TOU I-2 TOU II-3 |
IgA1 | I/II | Lefranc M-P. et al. (1983) [3] | |
| Italy | 4 | SAF I-2 DEM R.B. D.B. |
IgA1, IgG2, IgG4, IgE | III/III | Migone N. et al. (1984) [6] Bottaro A. et al. (1989) [7] Plebani A. et al. (1993) [8] |
| Tunisia | 1 | T17 | IgA1, IgG2, IgG4, IgE | III/III (2) | Wiebe V. et al. (1994) [5] |
| Sardinia | 1 | FRO I-2 | IgA1, IgG2, IgG4 | III/IV | Migone N. et al. (1984) [6] |
| Italy | 2 | TIM MOD |
IgG2 | III/del G2 | Bottaro A. et al. (1989) [7] |
| Italy | 1 | CRU | IgG2, IgG4, IgE, IgA2 | V/V | Bottaro A. et al. (1989) [7] |
| Sweden | 1 | NY (3) | IgG1 | VI/del G1 | Smith C.I.E. et al. (1989) [9] |
(1) The two grandsons of TAK3 display the same type of deletion, also in the homozygous state (Lefranc M-P and Lefranc G 1987) [10], Lefranc et al. (1991) [11].
(2) The T17 deletion was erroneously interpreted as of type IV in Chaabani et al. (1985) [11,12]. Wiebe et al. (1994) [5] demonstrate the deletion of the IGHE gene and the absence of IgE, and characterize the deletion as of type III.
(3) This individual has an increased susceptibility to infections, but there is no evidence that this was linked to the IGHC deletions. The IGHG1 deletion is most probably polymorphic as shows by Sukernik R.I. et al. (1992) [13] in populations of Northwestern Siberia.