The locus representation below
only shows the relative position of the IGH subgroups as deduced from deletion mapping analysis of murine pre-B and B cell lines
rearranged loci [4,6]. The number of genes per subgroup, estimated from Southern blot
hybridization studies and sequencing is according to Kofler et al. [5]
for the IGHV1-IGHV14 subgroups and to Mainville et al. [11] for the IGHV15 subgroup
[12].
As the murine IGH locus sequence has become publicly available, two detailed maps covering the C57BL/6 IGH locus
have recently been published [24]. These maps show the positions of 170 full-length IGHV genes, and of the IGHD,
IGHJ and IGHC genes from C57BL/6 (Ighb haplotype).
The new Locus representation: Mouse (Mus musculus) IGH was first presented at IMGC
(poster) and is available on the IMGT Web site since the 7th of November 2005.
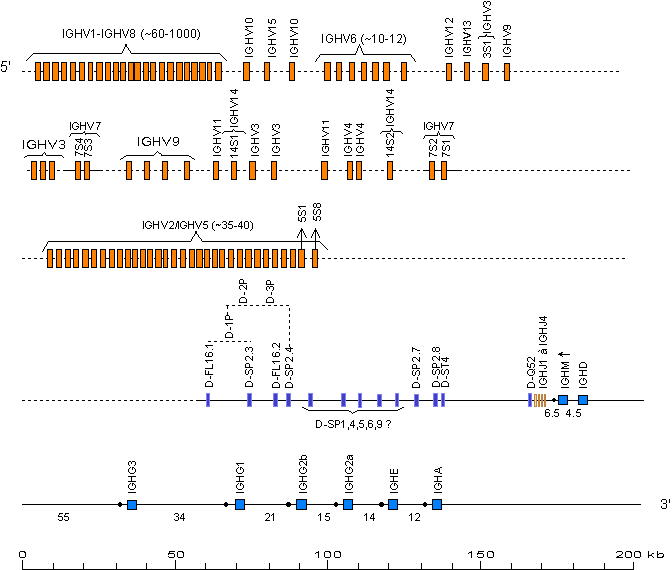
The boxes representing the genes are not to scale. Exons are not shown.
SWITCH sequences are represented by a
filled circle upstream of the IGHC genes.
- clan I: IGHV1, IGHV9, IGHV14 and IGHV15 subgroup genes
- clan II: IGHV2, IGHV3, IGHV8 and IGHV12 subgroup genes
- clan III: IGHV4, IGHV5, IGHV6, IGHV7, IGHV10, IGHV11 and IGHV13 subgroup genes
Legend:
Colors are according to IMGT color menu.
The boxes representing the genes are not to scale. Exons are not shown.
| The interspersed IGHV2 and IGHV5 subgroup genes are the most D-proximal genes [2]. | |
| IGHV3: | The IGHV3 subgroup genes map to 3 locations: three genes map 5' of the V7S4 and V7S3 genes V3S1 is the most 5' member of the IGHV3 subgroup. This gene is separated from the remainder 5' IGHV3 cluster by at least one IGHV9 gene [2]. |
| IGHV4: | The IGHV4 subgroup consists of two genes per haploid genome in BALB/c and C57BL/10. The IGHV4 subgroup genes maps 5' of the V7S1/V7S2 pair [2]. |
| IGHV6: | The IGHV6 subgroup maps 5' of the IGHV3 subgroup [2]. |
| IGHV7: | The IGHV7 subgroup consists of four genes per haploid genome in BALB/c and C57BL/10. Three are functional: V7S1, V7S3 and V7S4 and one is pseudogene: V7S2. The IGHV7 genes map as two distinct pairs: 5'-V7S2-V7S1-3' and 5'-V7S4-V7S3-3', the V7S2/V7S1 pair mapping more proximal to the D genes and nearest to the IGHV2/IGHV5 cluster [1,2]. |
| IGHV9: | The IGHV9 genes map to 2 distinct locations. The IGHV9 subgroup in BALB/c comprises a minimum of 6 genes [3,9]. |
| IGHV10: | The IGHV10 subgroup genes map 5' of the IGHV6 members and 3' of the most dowstream IGHV8 genes [8,11]. |
| IGHV13: | The IGHV13 subgroup, which may contain only one member, shares sequence similarity with the IGHV6 subgroup and its located to the 3' side of the IGHV6 subgroup genes with the IGHV12 gene being located between them. The IGHV13 gene has been mapped 5' to the most 5' IGHV3 gene (V3S1) [6,7]. |
| IGHV14: | The IGHV14 subgroup, with at least three genes, is related to the IGHV1 subgroup but is physically separated [7]. One gene V14S2 maps between the IGHV4 subgroup genes and the 3' pair V7S2/V7S1. The map position of the V14S1 is on the 5' side of the 3' IGHV3 subgroup cluster [10,11]. |
| IGHV15: | The two IGHV10 members are separated by the single IGHV15 gene [11]. |
For detailed references, see:
- IGHV genes
- [1] Crews, S. et al., Cell, 25, 59-66 (1981).
- [2] Brodeur, P.H. and Riblet, R., Eur. J. Immunol. 14, 922-930 (1984).
- [3] Winter, E. et al., EMBO J., 4, 2861-2867 (1985).
- [4] Brodeur, P.H. et al., J. Exp. Med., 168, 2261-2278 (1988).
- [5] Kofler, R. et al., J. Immunol., 140, 4031-4034 (1988).
- [6] Pennell, C.A. et al., Eur. J. Immunol., 19, 2115-2121 (1989).
- [7] Tutter, A. et al., J. Immunol., 147, 3215-3223 (1991).
- [8] Kofler, R. et al., Immunol. Rev., 128, 5-21 (1992).
- [9] Sims, M.J. et al., J. Immunol., 149, 1642-1648 (1992).
- [10] Sheehan, K.M. et al., J. Immunol., 151, 5364-5375 (1993).
- [11] Mainville, C.A. et al., J. Immunol., 156, 1058-1046 (1996).
- [12] Almagro, J.C. et al., Mol. Immunol., 34, 119-1214 (1997).
- IGHD genes
- [13] Kurosawa, Y. and Tonegawa, S., J. Exp. Med., 155, 201-218 (1982).
- [14] Woods, C. and Tonegawa, S., Proc. Natl. Acad. Sci. USA 80, 3030-3034 (1983).
- [15] Feeney, A.J and Ribbet, R., Immunogenetics, 37, 217-221 (1993).
- IGHJ genes
- [16] Early, P. et al., Cell, 19, 981-992 (1980).
- [17] Newell, N. et al., Science, 209, 1128-1132 (1980).
- [18] Sakano, H. et al., Nature, 286, 676-683 (1980).
- IGHC genes
- [19] Edelman, G.M. et al., Proc. Natl. Acad. Sci. USA, 63, 78-85 (1969).
- [20] Honjo, T. and Kataoka,T., Proc. Natl Acad. Sci. USA, 75, 2140-2144 (1978).
- [21] Honjo, T. et al., Immunological Rev., 59, 33-67 (1981).
- [22] Yamawaki-Kataoka, Y. et al., Proc. Natl. Acad. Sci. USA, 79, 2623-2627 (1982).
- [23] Shimiza, A. et al., Cell, 28, 499-506 (1982).
- IGH locus from the C57BL/6 genome sequencing
- [24] Riblet, R. In: Molecular Biology of B cells. Honjo, T., Alt, F.W. and Neuberger, M. eds, Elsevier Academic Press, London, pp 19-26 (2004).



