IMGT Repertoire (IG and TR)
Locus representation: human (Homo sapiens) IGK locus on chromosome 2 (2p11.2)
Lefranc, M.-P., Exp. Clin. Immunogenet., 18, 161-174 (2001). PMID:11549845
Lefranc, M.-P. and Lefranc, G., The Immunoglobulin FactsBook, Academic Press, London, 458 pages (2001).
Human (Homo sapiens) IGK locus on chromosome 2 (2p11.2)
The orientation of the human (Homo sapiens) IGK locus on chromosome 2 (2p11.2) is reverse (REV).
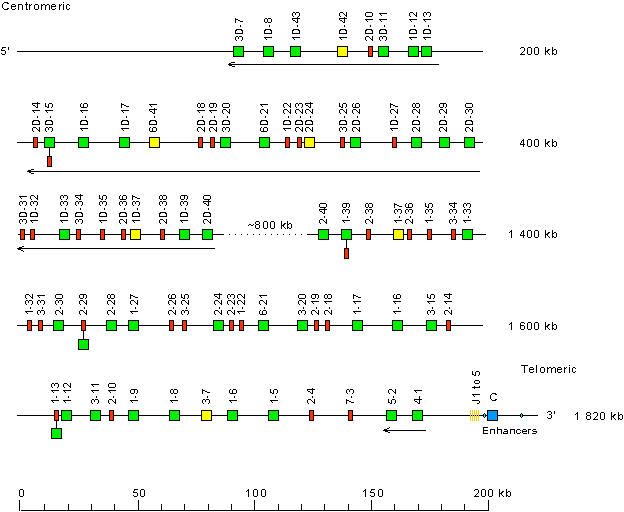
Legend:
Colors are according to IMGT color menu for genes.
The boxes representing the genes are not to scale. Exons are not shown.
A double slash // indicates a gap in genome assembly. Distances in bp (base pair) or in kb (kilobase) associated with a double slash are taken
into account in the length of the lines and included in the numbers displayed at the right end of the lines.
The IGKV genes of the proximal V-CLUSTER
are designated by a number for the subgroup, followed by a hyphen and a number for the localization from 3' to 5' in the locus.
The IGKV genes of the distal duplicated V-CLUSTER are designated by the same numbers as the corresponding
genes in the proximal V-CLUSTER, with the letter D added.
Arrows show the IGKV genes whose orientation is opposite to that of the
J-C-CLUSTER.
• No 5' borne conserved between species has been identified upstream of IGKV3D-7 (F), the most 5' gene in the locus.
• RPIA (ribose 5-phosphate isomerase A) (3' borne) has been identified 106 kb downstream of IGKC (F), the most 3' gene in the locus.
The Human (Homo sapiens) IGKV genes belong to seven subgroups (Gene table: Human IGKV) which are part of three clans (Figure IGKV clans). Clans comprise, respectively:
- clan I: IGKV1 subgroup genes
- clan II: IGKV2, IGKV3, IGKV4 and IGKV6 subgroup genes
- clan III: IGKV5 and IGKV7 subgroup genes
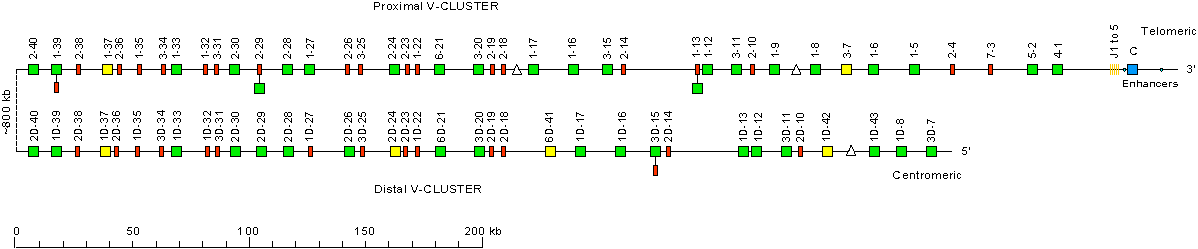
Alternative representation of the human (Homo sapiens) IGK locus on chromosome 2 (2p11.2)
This representation is taking into account the polymorphic duplication in the locus
(see references from Zachau's group below).
Small triangles indicate parts which are absent in the proximal or distal V-CLUSTER when considering
the duplication in the human IGK locus.
Locus representation with only the functional genes: human (Homo sapiens) IGK locus on chromosome 2 (2p11.2)
Gene localization is according to data from IMGT/LocusView.
This locus representation with only the functional genes is used with IMGT/GeneFrequency.
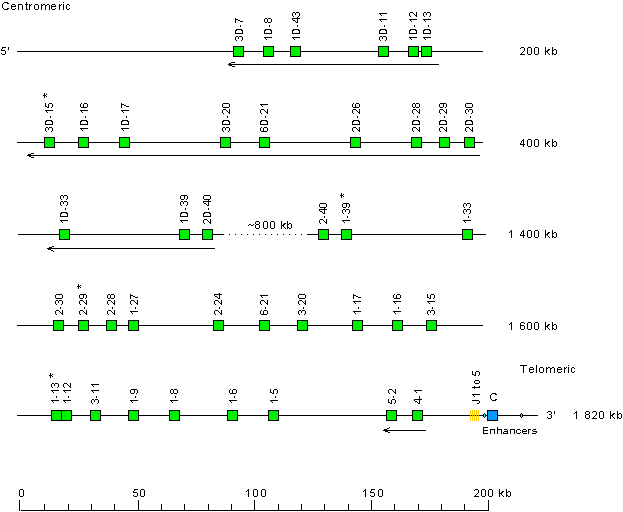
* When needed, indicate the presence of allele(s) with another functionality: see above human (Homo sapiens) IGK locus on chromosome 2 (2p11.2)
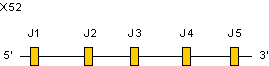
Zoom for IGKJ genes
- [1] Zachau, H.G., Gene, 135, 167-173 (1993).
- [2] Zachau, H.G., The Immunologist, 4, 49-54 (1996).
- [3] Lefranc, M.-P. and Lefranc, G., The immunoglobulin FactsBook, Academic Press, London, 458 pages (2001). 012441351X
- [4] Lefranc, M.-P., Nomenclature of the human immunoglobulin genes, Current Protocols in Immunology, A.1P.1-A.1P.37 (2000).
- [5] Hüber, C. et al., Eur. J. Immunol., 23, 2868-2875 (1993).
- [6] Schäble, K.F. and Zachau, H.G., Biol. Chem. Hoppe-Seyler, 374, 1001-1022 (1993).
- [7] Cox, J.P. et al., Eur. J. Immunol., 24, 827-836 (1994).
- [8] Schäble, K.F. et al., Biol. Chem. Hoppe-Seyler, 375, 189-199 (1994).
- [9] Hieter, P.A. et al., J. Biol. Chem., 257, 1516-1522 (1982).
- [10] Hieter, P.A. et al., Cell, 22,197-207 (1980).
- [11] Emorine, L. et al., Nature, 304, 447-449 (1983) (IGKJ-IGKC intron).
- [12] Gimble, J.M. and Max, E.E., Mol. Cell. Biol., 7, 15-25 (1987).
- [13] Judde, J.G. and Max, E.E., Mol. Cell. Biol., 12, 5206-5216 (1992).
- [14] Dunham, I. et al., Nature, 402, 489-495 (1999).
Chromosomal orphon set IGKV/OR22 on chromosome 22 (22q11)
The orientation of the human (Homo sapiens) IGK locus on chromosome 22 (22q11) is reverse (REV).
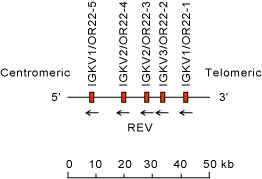
Legend:
Colors are according to IMGT color menu for genes.
The boxes representing the genes are not to scale. Exons are not shown.
A double slash // indicates a gap in genome assembly. Distances in bp (base pair) or in kb (kilobase) associated with a double slash are taken
into account in the length of the lines and included in the numbers displayed at the right end of the lines.
- Created:
- 07/10/1996
- Last updated:
- 22/09/2023
- Authors:
- Valérie Barbié and Marie-Paule Lefranc
