IMGT Repertoire (IG and TR)
Collier de Perles: channel catfish (Ictalurus punctatus)
IGHD1 CH2D (AF363448)
On one layer
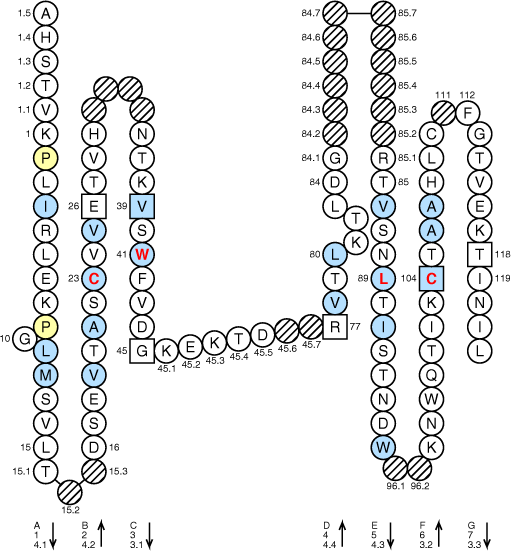
Colors are according to IMGT Color menu for sequence alignments and Color menu for CDR-IMGT
Legend:
- Amino acids are shown in the one-letter abbreviation.
- Hydrophobic amino acids (hydropathy index with positive value) and Tryptophan (W) found at a given position in more than 50% of analysed IG and TR sequences are shown in blue.
- All Proline (P) are shown in yellow.
- Missing positions in IMGT Colliers de Perles based on 3D structures may be due to incomplete experimental data.
- Arrows indicate the direction of the beta sheets and their different designations in 3D structures. This information has to be used carefully if not supported by experimental data.
- The positions 26, 39 and 104 are shown in squares by homology with the corresponding positions in the V-REGIONs. Positions 45 and 77 which delimit the characteristic "CD" strand of the C-DOMAINs, and position 118 which corresponds structurally to J-PHE or J-TRP of the J-REGION, are also shown in squares.
- Hatched circles or squares correspond to missing positions according to the IMGT unique numbering for C-DOMAINs
(Lefranc et al. 2005 PMID: 15572068
 ).
). - In green, position of amino acid changes involved in allotype polymorphism.
- Strands of the beta-pleated sheet localized at the interface between domains are shown in bold.
- The hinge six amino acids which belongs to the same exon as the CH2 domain are not shown.
- The CHS region is not shown.
See also:
- Created:
- 15/07/2003
- Last updated:
- Wednesday, 23-Oct-2024 09:53:41 CEST
- Authors:
- Nathalie Bosc
