
IMGT Repertoire (MH)
IMGT Collier de Perles for Oncorhynchus mykiss MH1-B (UAA) [D3] (C-LIKE-DOMAIN)
Oncmyk-UAA (AF091779)
Collier de Perles on one layer
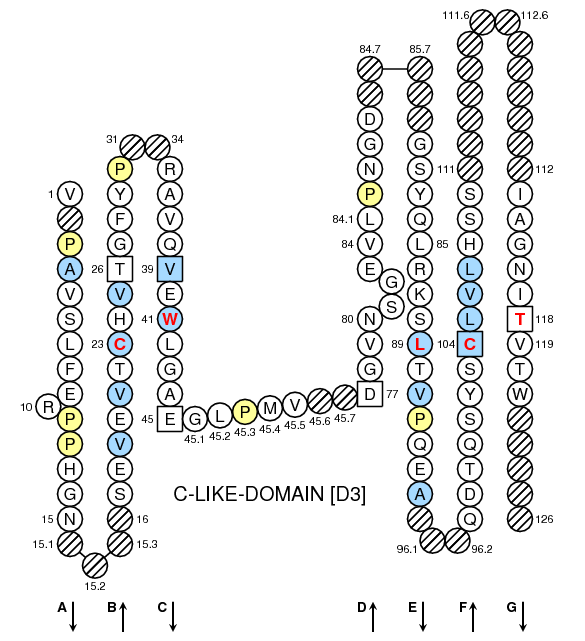
Collier de Perles on two layers
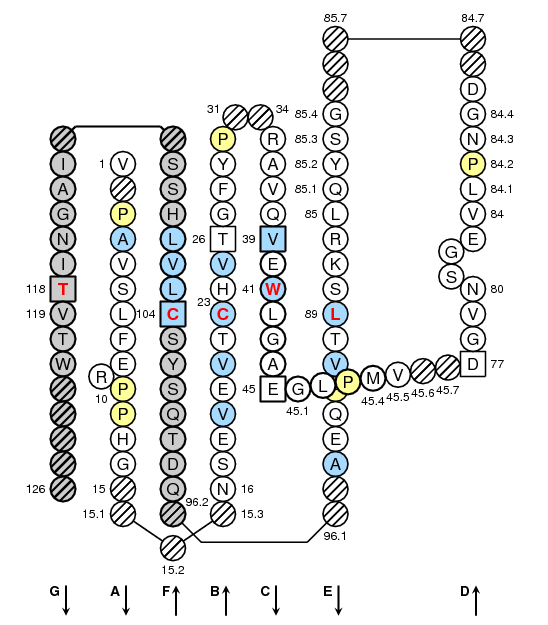
This figures have been generated with the IMGT/Collier de Perles tool for C-DOMAIN developed by Quentin Kaas (11/03/2002).
Legend:
- Amino acids are shown in the one-letter abbreviation.
- Hydrophobic amino acids (hydropathy index with positive value) and Tryptophan (W) found at a given position in more than 50 % of analysed Ig and TcR sequences are shown in blue.
- All Proline (P) are shown in yellow.
- Arrows indicate the direction of the beta sheets and their different designations in 3D structure. This information has to be used carefully if not supported by experimental data.
- Hachured circles or squares correspond to missing positions according to the IMGT unique numbering.
- Grey circles or squares correspond to missing positions according to the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN.
- Missing positions in IMGT Colliers de Perles based on 3D structures may be due to incomplete experimental data.
- The positions 26, 39 and 104 are shown in squares by homology with the corresponding positions in the V-REGIONs. Positions 45 and 77 which delimit the characteristic "CD" strand of the C-DOMAINs, and position 118 which corresponds structurally to J-PHE or J-TRP of the J-REGION, are also shown in squares.
See also:
- IMGT Repertoire:
- IMGT Scientific chart:
- Collier de Perles is described using the IMGT unique numbering for C-DOMAIN and C-LIKE-DOMAIN
- Rules for the gap and additional positions in C-DOMAIN and C-LIKE-DOMAIN
- IMGT color menu
- IMGT Index:
- IMGT Education (FR):
- Tools:


