IMGT® institutions 
IMGT®, the international ImMunoGeneTics information system®, http://www.imgt.org, created in 1989 by Marie-Paule Lefranc in Montpellier, France (Laboratoire d'ImmunoGénétique Moléculaire (LIGM), University of Montpellier (UM) and French National Center for Scientific Research (CNRS)) has for mission to be the global reference in immunogenetics and immunoinformatics.
IMGT® is in charge, since its creation, of the International Union of Immunological Societies (IUIS) Nomenclature SubCommittee, IMGT-NC, for the nomenclature of the genes and alleles of the immunoglobulins (IG) or antibodies and T cell receptors (TR) of all vertebrate species with jaws (gnasthostomata).
IMGT® is attached to CNRS, to the University of Montpellier and to the Ministry of Higher Education, Research and Innovation (MESRI).
IMGT® is a team of the Institut de Génétique Humaine (IGH), UMR 9002 CNRS-University of Montpellier.
Acknowledgments and Funding support
IMGT® attachment and affiliation 
International
- CNRS registered trademark (European Union, Canada, United States).
- The IMGT Nomenclature Committee (IMGT-NC) is the IUIS Immunoglobulins (IG), T cell Receptors (TR) and Major Histocompatibility (MH) Nomenclature Sub-Committee (IMGT-NC) since 1992.
- Academic Institutional Member of International Medical Informatics Association (IMIA) since 2006.
- Partner of the European Life sciences Infrastructure for Biological Information (ELIXIR) IFB French node.
- Member of the Global Alliance for the Genomics and Health (GA4GH) since 2015.
National
- Member of the Institut Français de Bioinformatique (IFB), since its creation in December 2014.
- Member of GDR CNRS n° 3003 Molecular Bioinformatics (BiM).
- Member of GDR CNRS n° 3260 Antibodies and Therapeutic targeting (ACCITH) (2009-2016).
- National Research Platform in Bioinformatics with the Research Inter-Organization label RIO (CEA, CNRS, INRA, INSERM), from the creation of RIO in 2001 and until the creation of the GIS IBiSA in 2007.
- Member of GIS Infrastructures in Health Biology and Agronomy (IBiSA), coordination of the platforms of Research in Living Sciences, since the creation of the GIS in 2007.
- Member of National Network of platforms in Bioinformatics (ReNaBi) (2007-2014).
Interregional
Regional
- CNRS, Délégation Régionale Languedoc-Roussillon, DR13.
- Université de Montpellier since the creation of IMGT® in 1989 (Plan Pluri-formation 1999-2009).
- Grand Plateau Technique for Research and Innovation Languedoc-Roussillon (GPTR), since the creation of the GPTR in 2005
- Bioinformatics platform of the Montpellier Languedoc-Roussillon Genopole®.
- Member of the GIS, Genopole Montpellier Languedoc-Roussillon (until 2007, creation of the GIS IBiSA).
Certifications
- HON Certification (since July 1999).
- Quality management systems: ISO 9001 (since October 2010), NFX 50-900, IQuaRe.
IMGT® authority
IMGT Director (since 2015): Sofia Kossida (Sofia.Kossida@igh.cnrs.fr), Professor, University of Montpellier
IMGT Founder and director (1989-2014): Marie-Paule
Lefranc
(Marie-Paule.Lefranc@igh.cnrs.fr),
Senior Member of the Institut Universitaire de France,
Professor Emeritus, University of Montpellier (2015-2019)
The Laboratoire d'ImmunoGénétique Moléculaire (LIGM) (Prof. Marie-Paule Lefranc and Prof. Gérard Lefranc), University of Montpellier, IGH (UMR 9002 CNRS-University of Montpellier), has been involved for more than 30 years in the genetics, structure, polymorphism and function of the immunoglobulins (IG) and T cell receptors (TR).
The scientific contribution of LIGM is shown by publications in international journals, communications, invitations to congresses and seminars, the isolation of 40 genomic probes. These probes, released by ATCC (American Type Culture Collection), JCRB (Japanese Cancer Research Resources Bank) and ICRF (Imperial Cancer Research Fund, Human Genetic Resource Centre), are used by many laboratories abroad, for fundamental research and clinical applications. Some of these probes are powerful tools in the characterisation of the B and T cells, the analysis of clonality in leukemias and lymphomas, and the therapeutics follow up (residual diseases). 134 genomic sequences have been sent to the EMBL/GenBank databases.
In 1989, LIGM started IMGT/LIGM-DB, the first specialised and integrated database on immunoglobulins (IG) and T cell receptors (TR). Since May 2000 and owing to the considerable expansion and success of IMGT®, the international ImMunoGeneTics information system®, the LIGM scientific activity has been mainly devoted to the IMGT® research and development.
IMGT® information system overview 
IMGT®, founded in Montpellier in 1989 by Marie-Paule Lefranc (Université Montpellier 2 and CNRS) is the international reference in immunogenetics and immunoinformatics. IMGT® is specialized in the sequences, genes and structures of immunoglobulins (IG) or antibodies, T cell receptors (TR), major histocompatibility (MH) proteins of vertebrates, IgSF and MhSF superfamily proteins of vertebrates and invertebrates, fusion proteins for immunological applications (FPIA) and composite proteins for clinical applications (CPCA).
IMGT® consists of databases, interactive tools and Web resources. IMGT® is used by researchers in academic and pharmaceutical laboratories in multiple areas of research:
- basic research,
- medical research (analysis of repertoires of antibody and T cell receptor recognition sites in normal (infections, cancers) and abnormal (autoimmune diseases, immunodeficiencies) immune responses and in proliferative syndromes (leukemias, lymphomas, myelomas)),
- veterinary research (study of the IG and TR repertoires in domestic and wild species),
- genomic research (study of the diversity and evolution of genes of the adaptive immune response),
- research in structural biology (evolution of IgSF and MhSF protein domains),
- biotechnology related projects of the Human Proteome Organisation (HUPO) and antibody engineering (single chain variable fragment scFv, Fab, combinatorial libraries, phage display, chimeric, humanized and human antibodies),
- diagnosis, prognosis and therapeutic monitoring of leukemias, lymphomas and myelomas (identification of the malignant clone(s) and assessment of residual diseases),
- therapeutic approaches (grafts, immunotherapy, vaccinology).
IMGT® is labeled Bioinformatics Platform RIO (2001), IBiSA (since 2007), Grand Plateau Technique for Research and Innovation Languedoc-Roussillon (since 2005), Cancéropôle Grand Sud-Ouest, ReNaBi, GDR CNRS: BiM, ACCITH, Labex MabImprove, Academic Institutional Member of the International Medical Informatics Association (IMIA) (since 2006). IMGT® is certified ISO 9001 (since 2010) and NFX 50-900 (since 2014).
- IMGT® 2003 (IMGT-Choreography)
- IMGT® 2004 (NAR 2005 figure)
- IMGT® 2007 (IMGT® databases and tools figure)
- IMGT® 2008 (IMGT® databases - ELIXIR survey 2008)
- IMGT® 2009 (Etude de cas du Ministère, in French, 53 pages, 16/11/09)
- IMGT® 2011 IGH Scientific report Aug 2011, p47-48
- IMGT® 2012 (IMGT® databases and tools figure)
- IMGT® 2012 IGH Scientific report Sept 1012, p47-48
- IMGT® 2012 Presentation at IGH days, Nov 2012 (diaporama)
- IMGT® 2013 IGH Scientific report Dec 2013, p49-50
- IMGT® 2014 IGH Scientific report Dec 2014, p45-46
- IMGT® 2015 IGH Department retreat Mèze
- IMGT® 2016 (IMGT® (in French, 12 pages) 27/01/16 - IFB web)
- IMGT® 2016 (IMGT® (in English 7 pages) 11/10/16 _ IFB core resource)
Databases
All databases are available at http://www.imgt.org
Sequence databases
- IMGT/LIGM-DB (LIGM, Montpellier, France)
- IMGT/MH-DB (ANRI, BPRC, hosted at EBI, Hinxton, UK)
- IMGT/PRIMER-DB (LIGM, Montpellier, EUROGENTEC, Belgium)
- IMGT/CLL-DB (LIGM, Montpellier, France)
Gene database
3D and 2D structure database
Monoclonal antibodies database
Interactive on-line tools
All interactive on-line tools are available at http://www.imgt.org
Sequence analysis
- IMGT/V-QUEST
- IMGT/HighV-QUEST
- IMGT/StatClonotype
- IMGT/JunctionAnalysis
- IMGT/Allele-Align
- IMGT/PhyloGene
- IMGT/DomainDisplay
Gene analysis
- IMGT/LocusView, IMGT/GeneView, IMGT/GeneSearch, IMGT/CloneSearch
- IMGT/GeneInfo (TIMC and ICH, Grenoble; LIGM, Montpellier)
- IMGT/GeneFrequency
2D and 3D structure analysis
Web resources
All IMGT Web resources are available at http://www.imgt.org
- IMGT Repertoire (locus representations, gene tables, allele alignments, 2D representations (or IMGT Colliers de Perles), 3D representations)
- IMGT Scientific chart (rules of the scientific chart)
- IMGT Index (a referential index)
- IMGT Bloc-notes (many links out)
- IMGT Education (educational pages)
IMGT® has been available on the Web since July 1995. IMGT® provides the biologists, researchers and clinicians with an easy to use and friendly interface. IMGT® has an exceptional response with more than 150,000 work sessions a month. Visitors are equally distributed between the European Union, the United States and the remaining world. IMGT/LIGM-DB data are also distributed by the HTTP or FTP. IMGT/LIGM-DB can be searched by BLAST or FASTA on different servers (EBI, Institut Pasteur, IGH...).
By its high quality and its easy data distribution, IMGT® has important implications in medical research (repertoires in autoimmune and infectious diseases, AIDS, leukemias, lymphomas, myelomas, allergy), veterinary research, functional genomics, genome diversity, genome evolution studies and comparative genomics of the adaptative immune system, biotechnology related to antibody engineering (combinatorial libraries, phage displays, single chain Fragment scFv, chimeric, humanized and human antibodies), diagnostic (detection and follow up of residual diseases), and therapeutical approaches (grafts, immunotherapy, vaccinology).
IMGT® has become the international reference in immunogenetics and immunoinformatics. At the cutting edge of immunology and bioinformatics, IMGT® is an invaluable integrated database for both fundamental, pharmaceutical, veterinary and clinical research.
IMGT®, the international ImMunoGeneTics information system® (IFB-ELIXIR Resource)
Resource description | Principal investigators | Year of establishment | Life cycle state | Overview | Access | Indicators | Translational stories | Funding | SAB | Updates | Recent citationsResource description
IMGT®, the international ImMunoGeneTics information system® (http://www.imgt.org) is the global reference in immunogenetics and
immunoinformatics, created in 1989 by Marie-Paule Lefranc (University of Montpellier and CNRS).
IMGT® is a high-quality integrated knowledge resource specialized in the immunoglobulins (IG) or
antibodies, T cell receptors (TR), major histocompatibility (MH) of human and other vertebrate species,
and in the immunoglobulin superfamily (IgSF), MH superfamily (MhSF) and related proteins of the immune
system (RPI) of vertebrates and invertebrates.
IMGT® provides a common access to sequence, genome and structure immunogenetics data, based on the
concepts of IMGT-ONTOLOGY and on the IMGT Scientific chart rules. IMGT® works in close collaboration
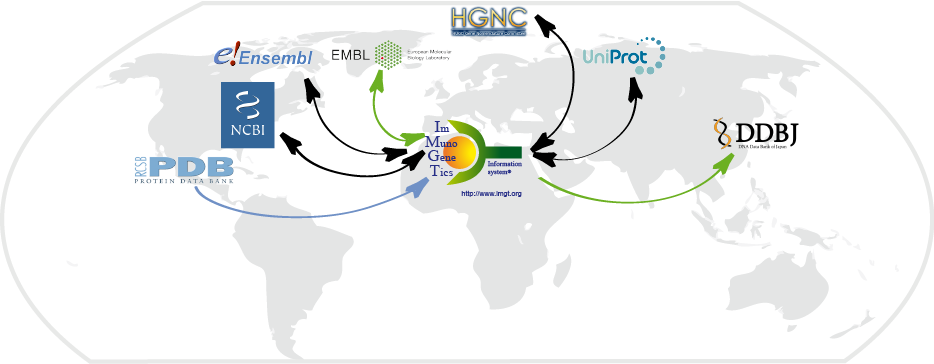
with EBI (Europe), DDBJ (Japan) and NCBI (USA).
More specifically, IMGT® uses the same accession numbers for nucleotide sequences as ENA, GenBank
and DDBJ. IMGT genes are officially approved by HUGO Nomenclature Committee (HGNC) since 1999 and used
by Ensembl (Hinxton, UK), NCBI Gene (which has direct links to IMGT/GENE-DB which is the international
reference database for IG and TR genes).
IMGT® consists of 7 databases (IMGT/LIGM-DB, IMGT/GENE-DB, etc) and 17 online
interactive tools for the analysis of sequences (IMGT/V-QUEST and its high throughput version
IMGT/HighV-QUEST for next generation sequencing (NGS), IMGT/DomainGapAlign, etc.), and the analysis of
genes and of three-dimensional (3D) structures. IMGT® also provides one standalone and more
than 20,000 pages of Web resources.
A schematic representation of the data exchanges is provided in the figure given below. IMGT®
resource is unique: there is no equivalent in the USA or elsewhere in the world.
Principal investigators:
Sofia KOSSIDA and Marie-Paule LEFRANCSofia KOSSIDA is IMGT® Director since 01/01/2015, Professor University of Montpellier. Marie-Paule LEFRANC is IMGT® Founder, IMGT® Director for 25 years (since IMGT® creation in 1989 up to 2014), Professor Emeritus University of Montpellier (2015-2019).
Year of establishment
1989Life cycle state :
Mature Stage: IMGT® is a mature ELIXIR Service. It is active and new data are currently integrated and reliable.Overview :
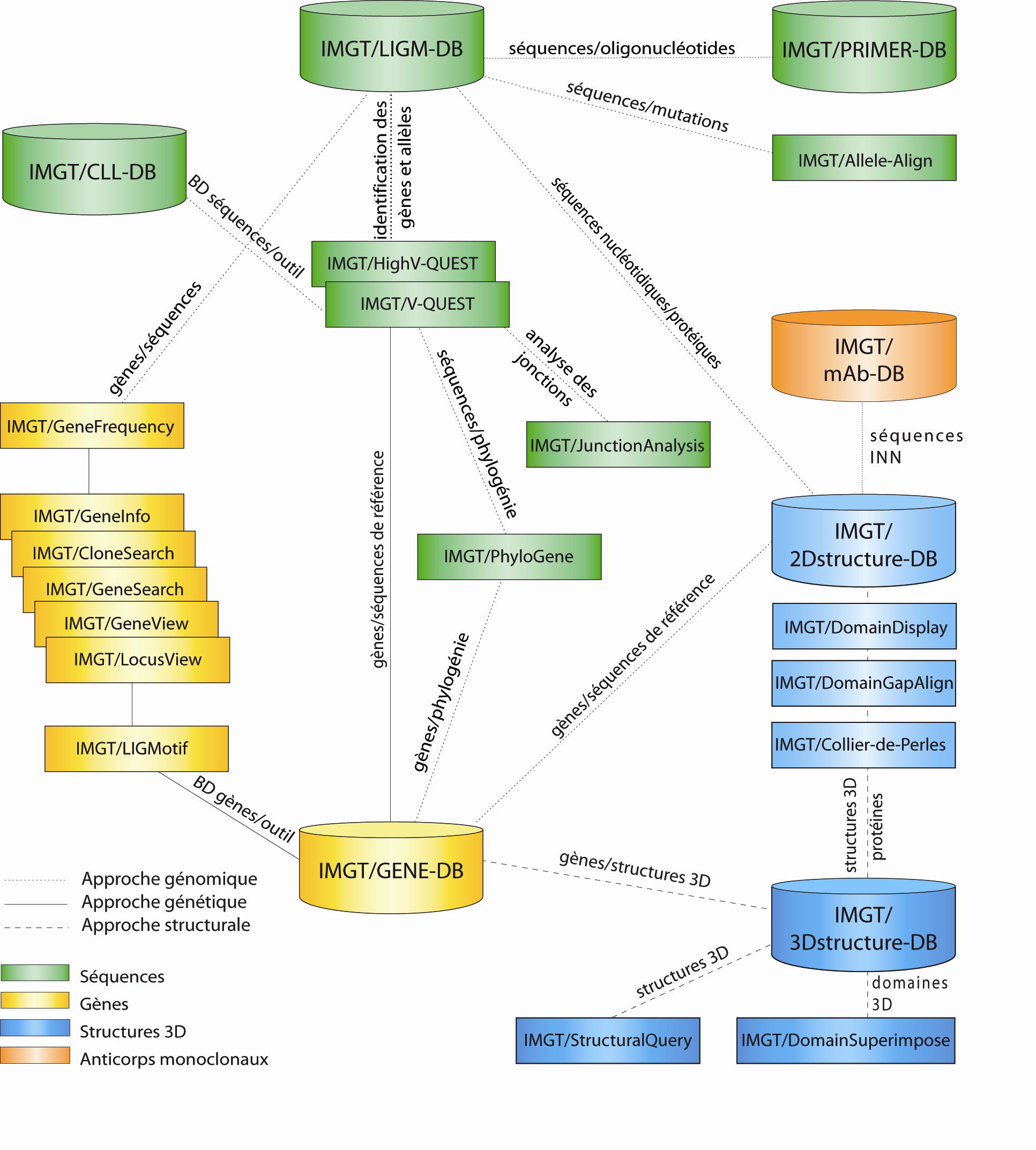
The IMGT®, the international ImMunoGeneTics information system® (http://www.imgt.org) IMGT® consists of 7 databases (cylinders) and 17 tools (rectangles). IMGT® also provides (not shown) IMGT/StatClonotype, a novel standalone tool which performs the statistical comparison of two sets of results from IMGT/HighV-QUEST for next generation sequencing (NGS) (1,000,000 sequences per batch) and more 20,000 pages de Web resources.
Access to the service/resource:
- Data provided by the French ELIXIR Node come from academic sources and are publicly available Data provided by IMGT® come from academic sources and are publicly available. Six IMGT databases are publicly available to academic users: IMGT/LIGM-DB (the IMGT® nucleotide database (196 440 sequences from 354 species in March 2020), IMGT/PRIMER-DB (the IMGT®primer database), IMGT/GENE-DB (the IMGT® gene database (7,759 genes and 10,069 alleles from 31 species, of which 723 genes and 1,629 alleles for Homo sapiens and 871 genes and 1,321 alleles for Mus musculus in March 2020), IMGT/2Dstructure-DB (for antibodies and other proteins for which the 3D structure is not available) and IMGT/3Dstructure-DB (for 3D structures, contact analysis and paratope/epitope interactions of IG/antigen and TR/peptide-MH complexes), IMGT/mAb-DB (interface for therapeutic antibodies and fusion proteins for immune applications (FPIA). IMGT/CLL-DB, requires a password as it is a working place for a consortium of clinicians and contains sequences from patients prior to submission to generalist databases (ENA, GenBank, DDBJ) and publications.
- Tools are distributed with a business-friendly open source license IMGT/StatClonotype, the IMGT® tool distributed as a standalone, is incorporated in the R package 'IMGTStatClonotype' under the LGPL licence. "The GNU Lesser General Public License (LGPL) is corporate-friendly and quite often used in R librairies. It allows for usage of certain library but modifications to it should be made public."
- Online services are freely accessible to users IMGT® online services are freely accessible to academic users. IMGT interactive tools accessible from the IMGT® Home page comprise: IMGT/V-QUEST (analysis of rearranged nucleotide sequence) and its high-throughput version IMGT/HighV-QUEST (1,000,000 nucleotide sequences per batch), IMGT/JunctionAnalysis, IMGT/Allele-Align, IMGT/PhyloGene, IMGT/DomainDisplay (amino acid sequences), IMGT/LocusView, IMGT/GeneView, IMGT/GeneSearch, IMGT/CloneSearch, IMGT/GeneInfo, IMGT/GeneFrequency, IMGT/DomainGapAlign (for amino acid sequence analysis of IG and TR variable and constant domains, and of MH groove domains), IMGT/Collier-de-Perles (for domain graphical 2D representation), IMGT/DomainSuperimpose, IMGT/StructuralQuery (for 3D structures). A password is required from the IMGT/HighV-QUEST users as this portal requires important computational resources for which IMGT® needs to apply twice a year (GENCI). All the IMGT® reference directories are publicly available online.
Indicators used to measure the quality and impact of service/resource
IMGT® uses the indicators of its Quality Management System. IMGT® has been approved by Lloyd's Register Quality Assurance France SAS to the Quality Management System Standards: ISO 9001:2008 since October, 18, 2010 and NFX 50-900 since October 13, 2014. "Research, development and provision of an integrated system (databases, tools and Web resources in immunogenetics and immunoinformatics". These indicators are analyzed regularly, by the Pilots of the SMQ processes and every year in January at the Steering Committee.Since the NFX certification, IMGT® has established Key Performance Indicators (KPI). These KPI have been reassessed at each steering committee since 2017 to take into account the NFX 50-900:2016 and ISO 9001:2015 (November 28, 2017).
Scientific focus and quality of science are measured by the number of publications, participations to meetings, invitations, new external collaborations and new contracts. Community served by the resource is measured by the annual survey (fundamental research, clinicians, biotechnology, big pharma, etc), e-mail exchanges, number of academic collaborations, educational workshops (IG-CLL), networks (EuroClonality), participation to education abroad on antibody informatics (USTH Hanoi, Vietnam), permanent education (Biocampus).
Quality of service is measured by the SMQ indicators (e.g., response time to user messages, survey).
Translational stories
IMGT® is in charge, since its creation in 1989 (HGM10, New Haven), of providing the international immunoglobulin (IG) and T cell receptor (TR) gene nomenclature for vertebrate species from fishes to humans.
By its creation in 1989, IMGT® marked the advent of immunoinformatics, which emerged at the
interface between immunogenetics and bioinformatics. For the first time, immunoglobulin (IG) or antibody
and T cell receptor (TR) variable (V), diversity (D), joining (J), and constant (C) genes were
officially recognized as "genes" as well as the conventional genes. This major breakthrough
allowed genes and data of the complex and highly diversified adaptive immune responses to be managed in
genomic databases and tools.
The IMGT® Homo sapiens IG and TR gene names were approved by the Human Genome Organisation
(HUGO) Nomenclature Committee (HGNC) in 1999 and were endorsed by the WHO-IUIS Nomenclature Subcommittee
for IG and TR. The IMGT® IG and TR gene names are the official international reference and, as such,
have been entered in IMGT/GENE-DB, in the Genome Database (GDB), in LocusLink at the National Center for
Biotechnology Information (NCBI) USA, in Entrez Gene (NCBI) when this database (now designated as
"Gene") superseded LocusLink, in NCBI MapViewer, in Ensembl at the European Bioinformatics
Institute (EBI), and in the Vertebrate Genome Annotation (Vega) Browser at the Wellcome Trust Sanger
Institute (UK). HGNC, Gene NCBI, Ensembl, and Vega have direct links to IMGT/GENE-DB.
IMGT® human IG and TR genes were also integrated in IMGT-ONTOLOGY on the NCBO BioPortal and, on the
same site, in the HUGO ontology and in the National Cancer Institute (NCI) Metathesaurus. Since 2007,
IMGT® gene and allele names have been used for the description of the therapeutic mAb and FPIA of
the WHO INN Programme. Amino acid sequences of human IG and TR constant genes (e.g., Homo
sapiens IGHM, IGHG1, IGHG2) were provided to UniProt in 2008. The current collaboration has for
aim the entry of the Homo sapiens germline IG variable genes in UniProt.
As an ELIXIR resource, IMGT® will actively pursue the current collaborations with the different
genome databases of species newly sequenced and the laboratories engaged in the characterization of the
IG and TR genes (UK, Italy, USA, etc.). Indeed the same IMGT concepts IDENTIFICATION (standardized
keywords), DESCRIPTION (labels), CLASSIFICATION (gene and allele nomenclature), NUMEROTATION (IMGT
unique numbering) and IMGT Scientific chart rules are used whatever the species from fishes to humans.
IMGT® high-quality and integrated system provides the resource for the standardized analysis of the expressed IG and TR repertoire of the adaptive immune responses in humans and other vertebrate species (e.g., animal models). They are used in basic, veterinary, and medical research, and are at the forefront of major methodological advances and medical implications. Examples of clinical applications (mutation analysis in leukemia and lymphoma, NGS repertoire analysis of the adaptive immune responses) and of pharmaceutical research and biotechnology (therapeutic antibody engineering and humanization) are given below.
- Mutation analysis in leukemia and lymphoma
- NGS repertoire analysis of the adaptive immune responses
- More than 21 billions of sequences analyzed to date (March 2020)
- 3203 users from 46 countries (43% from USA, 34% from EU, 23% from the remaining world) (March 2020)
IMGT/V-QUEST is frequently used by clinicians for the analysis of IG somatic hypermutations in leukemia, lymphoma, and myeloma, and more particularly in chronic lymphocytic leukemia (CLL) in which a low percentage of mutations of the rearranged IGHV gene in the VH of the leukemic clone has a poor prognostic value for the patients. For this evaluation, IMGT/V-QUEST is the standard recommended by the European Research Initiative on CLL (ERIC) for comparative analysis between laboratories. The sequences of the V-(D)-J junctions determined by IMGT/JunctionAnalysis are also used in the characterization of stereotypic patterns in CLL and for the synthesis of probes specific of the junction for the detection and follow-up of minimal residual diseases (MRD) in leukemias and lymphomas. A new era is opening up in hemato-oncology with the use of NGS for analysis of the clonality and MRD identification, making IMGT® standards usage more needed as ever.
IMGT/HighV-QUEST is the first web portal for next generation sequencing (NGS) immunoprofiling (online since October 2010). It is a paradigm for identification of IMGT clonotype diversity and standardized comparison of the NGS IG and TR repertoire analysis (diversity and expression) of the adaptive immune responses in normal (vaccination) and pathological situations (infectious diseases, cancers).
The therapeutic monoclonal antibody engineering field represents the most promising potential in medicine. IMGT® standards usage is needed more than ever before in antibody humanization and engineering, IG and TR engineering for immunotherapy, search of new specificities and targets, paratope/epitope characterization as demonstrated by the IMGT/DomainGapAlign tool and the IMGT/2Dstructure-DB, IMGT/3Dstructure-DB and IMGT/mAb-DB databases. Indeed, a standardized analysis of IG genomic and expressed sequences, structures, and interactions is crucial for a better molecular understanding and comparison of the mAb specificity, affinity, half-life, Fc effector properties, and potential immunogenicity. IMGT-ONTOLOGY concepts have become a necessity for IG loci description of newly sequenced genomes, antibody structure/function characterization, antibody engineering [single chain Fragment variable (scFv), phage displays, combinatorial libraries] and antibody humanization (chimeric, humanized, and human antibodies). IMGT® standardization allows repertoire analysis and antibody humanization studies to move to novel high-throughput methodologies with the same high-quality criteria. The CDR-IMGT lengths are now required for mAb INN applications and are included in the WHO INN definitions, bringing a new level of standardized information in the comparative analysis of therapeutic antibodies.
Funding of the service/resource
(1) IMGT® is a team of the Institut de Génétique Humaine (IGH, UMR 9002 CNRS-University of Montpellier). The permanent positions depend on the French Ministère de la Recherche et de l'Enseignement Supérieur. The present and former IMGT® directors are Professors of the University of Montpellier. There are 5 permanent positions: 2 from the University (1 PU, 1 research engineer IR) and 3 from CNRS (1 research engineer IR, 2 engineers IE).
(2) The salaries of the 8-10 CDD IE CNRS (biocurators, bioinformaticians) (turn-over of 3 years) are financed on pharmaceutical industry agreements with CNRS.
(3) Biocampus, Unité Mixte de Service (UMS 3426 CNRS - US 09 INSERM - UM), contributes to the renewal of PC and/or servers.
(4) IMGT/HighV-QUEST is granted access to the HPC@LR and to the High Performance Computing (HPC) resources of the Centre Informatique National de l'Enseignement Supérieur (CINES) and to Très Grand Centre de Calcul (TGCC) of the Commissariat l'Energie Atomique et aux Energies Alternatives (CEA) under the allocation [036029] (2010-2020) made by GENCI (Grand Equipement National de Calcul Intensif).
Previous funding sources include several EU grants (BIOMED, BIOTECH, 5th PCRDT, 6th PCRDT), IBiSA, ANR and Region Languedoc-Roussillon funding (listed on the web site).
SAB evaluating the resource
IMGT® has an international Scientific Committee since 1994. IMGT® is regularly evaluated scientifically by the SAB of the IGH (UMR 9002 CNRS-University of Montpellier).
Updates
IMGT® databases, tools and Web resources are constantly updated. New version numbers and dates are indicated on the web site and the users have the possibility to subscribe to the IMGT news RSS to be informed.
Recent citations (over the last three years)
Pégorier P., Bertignac M., Chentli I., Nguefack Ngoune V., Folch G., Jabado-Michaloud J., Hadi-Saljoqi S., Giudicelli V., Duroux P., Lefranc M.-P. and Kossida S. IMGT® Biocuration and Comparative Study of the T Cell Receptor Beta Locus of Veterinary Species Based on Homo sapiens TRB. Front. Immunol., 05 May 2020 doi: 10.3389/fimmu.2020.00821. PMID: 32431713 Radtanakatikanon A., Keller S.M., Darzentas N., Moore P.F., Folch G., Nguefack Ngoune V., Lefranc M.-P., Vernau W. Topology and expressed repertoire of the Felis catus T cell receptor loci. BMC Genomics. 2020 Jan 6;21(1):20. doi: 10.1186/s12864-019-6431-5. PMID: 31906850 Magadan S., Krasnov A., Hadi Saljoqi S., Afanasyev S., Mondot S., Lallias D., Castro R., Salinas I., Sunyer O., Hansen J., Koop B.F., Lefranc M-P., Boudinot P. Standardized IMGT® nomenclature of Salmonidae IGH genes, the paradigm of Atlantic salmon and rainbow trout: from genomics to repertoires. Front. Immunol., 12 November 2019 doi: 10.3389/fimmu.2019.02541. PMID: 31798572 Mondot S., Lantz O., Lefranc M.-P., Boudinot P. The T cell receptor (TRA) locus in the rabbit (Oryctolagus cuniculus): Genomic features and consequences for invariant T cells. Eur J Immunol. 2019 Jul 29. doi: 10.1002/eji.201948228. [Epub ahead of print]. PMID: 31355919 Lefranc M-P. Lefranc G. IMGT® and 30 years of Immunoinformatics Insight in Antibody V and C Domain Structure and Function. In Jefferis R; Strohl W. R., Kato K. Antibodies (Basel). 2019 Apr 11;8(2) 29. pii: E29. doi: 10.3390/antib8020029. PMID: 31544835 Ohlin M., Scheepers C., Corcoran M., Lees W.D., Busse C.E., Bagnara D., Thörnqvist L., Bürckert J.-P., Jackson K.J.L., Ralph D., Schramm C.A., Marthandan N., Breden F., Scott J., Matsen IV F.A., Greiff V., Yaari G., Kleinstein S.H., Christley S., Sherkow J.S., Kossida S., Lefranc M.-P., van Zelm M.C., Watson C.T., Collins A.M. Inferred allelic variants of immunoglobulin receptor genes: a system for their evaluation, documentation, and naming. Front Immunol. 2019 March 18. doi.org/10.3389/fimmu.2019.00435 Open access. PMID: 30936866 2018 Xochelli A, Bikos V, Polychronidou E, Galigalidou C, Agathangelidis A, Charlotte F, Moschonas P, Davis Z, Colombo M, Roumelioti M, Sutton L-A, Groenen P, van den Brand M, Boudjoghra M, Algara P, Traverse-Glehen A, Ferrer A, Stalika E, Karypidou M, Kanellis G, Kalpadakis C, Mollejo M, Pangalis G, Vlamos P, Amini R-M, Pospisilova S, Gonzalez D, Ponzoni M, Anagnostopoulos A, Giudicelli V, Lefranc M-P, Espinet B, Panagiotidis P, Piris MA, Du M-Q, Rosenquist R, Papadaki T, Belessi C, Ferrarini M, Oscier D, Tzovaras D, Ghia P, Davi F, Hadzidimitriou A, Stamatopoulos K. Disease-biased and shared characteristics of the immunoglobulin gene repertoires in marginal zone B cell lymphoproliferations. J Pathol. 2018 Nov 28. doi: 10.1002/path.5209. [Epub ahead of print]. PMID: 30484876 Lefranc M-P, Ehrenmann F., Kossida S., Giudicelli V., Duroux P. Use of IMGT® databases and tools for antibody engineering and humanization. In Nevoltris D and Chames P. (Eds) Antibody engineering, Humana Press, Springer, New York, USA. Methods Mol Biol. 2018;1827:35-69. doi: 10.1007/978-1-4939-8648-4_3. PMID: 30196491 Bradbury ARM, Trinklein ND, Thie H, Wilkinson IC, Tandon AK, Anderson S, Bladen CL, Jones B, Aldred SF, Bestagno M, Burrone O, Maynard J, Ferrara F, Trimmer JS, Görnemann J, Glanville J, Wolf P, Frenzel A, Wong J, Koh XY, Eng HY, Lane D, Lefranc M-P, Clark M, Dübel S. When monoclonal antibodies are not monospecific: hybridomas frequently express additional functional variable regions MAbs. 2018 May/Jun;10(4):539-546. doi: 10.1080/19420862.2018.1445456. Epub 2018 Mar 29. PMID: 29485921 Free PMC Article. Han SY, Antoine A, Howard D, Chang B, Chang WS, Slein M, Deikus G, Kossida S, Duroux P, Lefranc M-P, Sebra RP, Smith ML, Fofana IBF. Coupling of single molecule, long read sequencing with IMGT/HighV-QUEST analysis expedites identification of SIV gp140-specific antibodies from scFv phage display libraries. Front Immunol. 2018 Mar 1;9:329. doi: 10.3389/fimmu.2018.00329 Open access. PMID: 29545792 Martin J, Ponstingl H, Lefranc M-P, Archer J, Sargan D, Bradley A. Comprehensive annotation and evolutionary insights into the canine (Canis lupus familiaris) antigen receptor loci. Immunogenetics. 2018 Apr; 70(4):223-236. doi: 10.1007/s00251-017-1028-0. Epub 2017 Sep 19. PMID: 28924718IMGT® databases and tools (ELIXIR) 
Home page: http://www.imgt.org
Interface: Web UI
Affiliation: IMGT
Collection: IMGT databases and tools
| Name | Description | Function | Topic | Contact | Resource type | Tag | Publications | |
|---|---|---|---|---|---|---|---|---|
|
I M G T ® d a t a b a s e s |
IMGT/LIGM-DB | The IMGT® database for nucleotide sequences with translation of immunoglobulins (IG)
and T cell receptors (TR). Annotation is based on the IMGT-ONTOLOGY concepts. |
Query
and retrieval -
Input: ID Output: Flat file, annotation report (HTML) |
Immunoinformatics, Immunogenetics, Nucleotide sequences | Joumana Jabado-Michaloud Joumana.Michaloud@igh.cnrs.fr Data Patrice Duroux Patrice.Duroux@igh.cnrs.fr Developer |
Database | - Immunoglobulin (IG) - Antibody - T cell receptor (TR) |
PMID: 16381979 PMID: 25378316 |
| IMGT/PRIMER-DB | The IMGT® database for oligonucleotides (primers) of immunoglobulins (IG) and T cell
receptors (TR). Annotation is based on the IMGT-ONTOLOGY concepts. |
Query
and retrieval - Input : ID Output : Entry cards (HTML) |
Immunoinformatics, Immunogenetics, Nucleotide sequences | Geraldine Folch Geraldine.Folch@igh.cnrs.fr Data Patrice Duroux Patrice.Duroux@igh.cnrs.fr Developer |
Database | - Immunoglobulin (IG)
- Antibody - T cell receptor (TR) |
PMID: 15089751 PMID: 15972004 PMID: 25378316 |
|
| IMGT/GENE-DB | The IMGT® database for immunoglobulin (IG) and T cell receptor (TR) genes and alleles
(international nomenclature). Annotation is based on the IMGT-ONTOLOGY concepts. |
Query
and retrieval - Input : ID Output : Entry cards (HTML) |
Immunoinformatics, Immunogenetics, Genes and alleles | Véronique Giudicelli Veronique.Giudicelli@igh.cnrs.fr | Database | - Immunoglobulin (IG)
- Antibody - T cell receptor (TR) |
PMID: 15608191 PMID: 25378316 |
|
| IMGT/3Dstructure-DB | The IMGT® database for 3D structures of immunoglobulins (IG) or antibodies, T cell
receptors (TR), major histocompatibility (MH) proteins, related proteins of the immune
system (RPI) and fusion proteins for immune applications (FPIA). Annotation is based on the IMGT-ONTOLOGY concepts. |
Query
and retrieval - Input : ID Output : Entry cards (HTML) |
Immunoinformatics, Immunogenetics, Structures, Contact analysis | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Database | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) - Immunoglobulin superfamily (IgSF) - Major histocompatibility (MhSF) |
PMID: 14681396 PMID: 19900967 PMID: 21632774 PMID: 25378316 |
|
| IMGT/2Dstructure-DB | The IMGT® database for 2D structures (IMGT Colliers de Perles) of immunoglobulins (IG)
or antibodies, T cell receptors (TR), major histocompatibility (MH) proteins and related
proteins of the immune system (RPI). Annotation is based on the IMGT-ONTOLOGY concepts. |
Query
and retrieval - Input : ID Output : Entry cards (HTML) |
Immunoinformatics, Immunogenetics Amino acid sequences | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Database | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) - Immunoglobulin superfamily (IgSF) - Major histocompatibility (MhSF) |
PMID: 25378316 | |
| IMGT/mAb-DB | The IMGT® database for monoclonal antibodies (mAb) or immunoglobulins (IG), fusion
proteins for immune applications (FPIA) and composite proteins for clinical applications
(CPCA). IMGT/mAb-DB provides links to IMGT/2Dstructure-DB and IMGT/3Dstructure-DB. |
Query
and retrieval - Input : ID Output : Entry cards (HTML) |
IMGT-ONTOLOGYand Immunoinformatics, Immunogenetics, IG, antibodies, FPIA, CPCA, Specificity | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Database | - Immunoglobulin (IG) or antibody
- Fusion proteins for immune applications (FPIA) - Composite proteins for clinical applications (CPCA) |
PMID: 25378316 | |
|
I M G T ® o n l i n e t o o l s |
IMGT/V-QUEST | IMGT® tool for nucleotide sequence alignment and analysis of immunoglobulin (IG) or
antibody and T cell receptor (TR) variable domains, integrates IMGT/JunctionAnalysis,
IMGT/Automat and IMGT/Collier-de-Perles. Analysis is based on the IMGT-ONTOLOGY concepts. |
Nucleotide sequence analysis Input : FASTA Output: Report (HTML, Excel) |
Immunoinformatics, Immunogenetics, IG, TR, Nucleotide sequence analysis, V domain, V-(D)-J-region | Véronique Giudicelli Veronique.Giudicelli@igh.cnrs.fr |
Tool (analysis) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) |
PMID:15215425 PMID: 18503082, PMID: 21632778, PMID: 25378316 |
| IMGT/HighV-QUEST | IMGT® portal for NGS high-throughput nucleotide sequence analysis of immunoglobulins
(IG) and T cell receptors (TR) variable domains, integrates IMGT/JunctionAnalysis and
IMGT/Automat. Analysis is based on the IMGT-ONTOLOGY concepts. |
Next Generation Sequencing (NGS) nucleotide sequence analysis
Input : FASTA Output: Files (pdf, csv) |
Immunoinformatics, Immunogenetics, NGS nucleotide sequence analysis | Véronique Giudicelli Veronique.Giudicelli@igh.cnrs.fr Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (analysis) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) |
Immunome Research 2012, 8:1:2,
PMID: 23995877 PMID: 25378316 |
|
| IMGT/JunctionAnalysis | IMGT® tool for the analysis of the nucleotide sequences of the V-J and V-D-J junctions
of the variable domains of the immunoglobulins (IG) or antibodies and T cell receptors
(TR). Analysis is based on the IMGT-ONTOLOGY concepts. |
Nucleotide sequence analysis Input: FASTA Output: Report (HTML) |
Immunoinformatics, Immunogenetics, Nucleotide sequence analysis | Véronique Giudicelli Veronique.Giudicelli@igh.cnrs.fr |
Tool (analysis) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) |
PMID: 15262823 PMID: 21632777 PMID: 25378316 |
|
| IMGT/Allele-Align | IMGT® tool for the comparison of two alleles highlighting the nucleotide and amino acid differences. | Nucleotide sequence alignment Input: FASTA Output: Display (HTML) |
Sequence alignment, Alleles, Nucleotide sequence analysis | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (analysis) | PMID: 15089751 PMID: 15972004 PMID: 25378316 |
||
| IMGT/PhyloGene | IMGT® tool to compute and draw phylogenetic trees for immunoglobulin (IG) and T cell
receptor (TR) V-REGION nucleotide sequences. Analysis is based on the IMGT-ONTOLOGY concepts. |
Nucleotide sequence alignment
and phylogeny trees Input: FASTA Output: Display (HTML) |
Immunoinformatics, Immunogenetics, Gene families, Phylogeny | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (analysis) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) |
PMID: 12818634 PMID: 25378316 |
|
| IMGT/DomainDisplay | The IMGT® tool for the display of the amino acid sequences of the variable (V),
constant (C) and groove (G) domains. Analysis is based on the IMGT-ONTOLOGY concepts. |
Amino acid sequence analysis Input: FASTA Output: Display (HTML) |
Immunoinformatics, Immunogenetics, Amino acid sequences, Domain | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (analysis) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) - Immunoglobulin superfamily (IgSF) - Major histocompatibility (MhSF) |
PMID: 25378316 | |
| IMGT/LocusView | IMGT® tool providing a view of immunoglobulin (IG), T cell receptor (TR) and major
histocompatibility (MH) loci. Display is based on the IMGT-ONTOLOGY concepts. |
Query
and retrieval - Input : locus name Output: Display (HTML) |
Immunoinformatics, Immunogenetics, | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (visualiser) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) |
PMID: 15089751 PMID: 15972004 PMID: 25972004 |
|
| IMGT/GeneView | IMGT® tool providing the view of a gene in IMGT/LocusView. Display is based on the IMGT-ONTOLOGY concepts. | Query
and retrieval - Input : gene name Output: Display (HTML) |
Immunoinformatics, Immunogenetics, | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (query and retrieval) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) |
PMID: 15089751 PMID: 15972004 PMID: 25378316 |
|
| IMGT/GeneSearch | IMGT® tool allowing the search of a gene in IMGT/LocusView. Search is based on the IMGT-ONTOLOGY concepts. | Query
and retrieval - Input : gene name Output: Report (HTML) |
Immunoinformatics, Immunogenetics, IG, TR, MH, Gene names | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (query and retrieval) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) |
PMID: 15089751 PMID: 15972004 PMID: 25378316 |
|
| IMGT/CloneSearch | IMGT® tool allowing the search of a clone in IMGT/LocusView. Search is based on the IMGT-ONTOLOGY concepts. | Query
and retrieval - Input: clone name Output: Report (HTML) |
Immunoinformatics, Immunogenetics | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (query and retrieval) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) |
PMID: 15089751 PMID: 15972004 PMID: 25378316 |
|
| IMGT/GeneInfo | IMGT® tool providing combination (V-J and V-V) information for the human and mouse T cell receptor (TR) loci. Analysis is based on the IMGT-ONTOLOGY concepts. | Query and retrieval - | Immunoinformatics, Immunogenetics, Nucleotide sequences | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (visualiser) | - T cell receptor (TR) | PMID: 14681357 PMID: 16640788 PMID: 25378316 |
|
| IMGT/GeneFrequency | IMGT® tool providing the frequency usage of immunoglobulin (IG) and T cell receptor
(TR) variable (V), diversity (D) and joining (J) genes in Homo sapiens and Mus musculus
rearranged sequences from IMGT/LIGM-DB. Analysis is based on the IMGT-ONTOLOGY concepts. |
Query
and retrieval - Input: sequences from IMGT/LIGM-DB Output: Bar graphs of gene usage (V, D or J). |
Immunoinformatics, Immunogenetics Gene names | Patrice Duroux Patrice.Duroux@igh.cnrs.fr | Tool (visualiser) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) |
PMID: 15972004 PMID: 16640788 PMID: 25378316 |
|
| IMGT/DomainGapAlign | IMGT® tool for the analysis of amino acid sequences of variable (V), constant (C) or groove (G) domains. Analysis is based on the IMGT-ONTOLOGY concepts. | Amino acid sequence analysis
Input: FASTA Output: Display of aligned V, C or G domain sequences, gapped according to the IMGT unique numbering |
Immunoinformatics, Immunogenetics, Amino acid sequences Gene families Domain | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (analysis) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) - Immunoglobulin superfamily (IgSF) - Major histocompatibility (MhSF) |
PMID: 19900967 PMID: 21632775 PMID: 25378316 |
|
| IMGT/Collier-de-Perles | IMGT® tool to draw standardized IMGT 2D graphical representations, or IMGT Colliers de Perles, of variable (V), constant (C), groove (G) domains, starting from the user own domain amino acid sequences. IMGT Colliers de Perles are based on amino acid sequences gapped according to the IMGT-ONTOLOGY concept of IMGT unique numbering. | Graphical representation or IMGT Colliers de Perles of V, C or G domain Input: Amino acid sequences, gapped based on the IMGT unique numbering Output: IMGT Collier de Perles |
Immunoinformatics, Immunogenetics, Amino acid sequence Gene families Domain | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (visualiser) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) - Immunoglobulin superfamily (IgSF) - Major histocompatibility superfamily (MhSF) |
PMID: 11862387 PMID: 18208865 PMID: 21632776 PMID: 25378316 |
|
| IMGT/DomainSuperimpose | IMGT® tool to superimpose 3D structures of two domains from IMGT/3Dstructure-DB. Domain 3D structures are numbered according to the IMGT-ONTOLOGY concept of IMGT unique numbering. | Domain 3D structure comparison
Input: 3D structure domain ID
Output: Display of the superimposed 3D structures |
Immunoinformatics, Immunogenetics Domain | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (visualiser) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Major histocompatibility (MH) - Immunoglobulin superfamily (IgSF) - Major histocompatibility superfamily (MhSF) |
PMID: 25378316 | |
| IMGT/StructuralQuery | IMGT® tool to retrieve IMGT/3Dstructure-DB entries using amino acid structural
criteria for variable (V), constant (C) or groove (G) domains. Annotation is based on the IMGT-ONTOLOGY concepts. |
Query and retrieval - | Immunoinformatics, Immunogenetics, Domain | Patrice Duroux Patrice.Duroux@igh.cnrs.fr |
Tool (query and retrieval) | - Immunoglobulin (IG) or antibody
- T cell receptor (TR) - Immunoglobulin superfamily (IgSF) |
PMID: 15089751 PMID: 14681396 PMID: 25378316 |
IMGT®, the international ImMunoGeneTics information system® (IFB-ReNaBi) 
Information | Infrastructure | Données | Outils | Expertise |
Formations | Ouverture | Publications
Information
ContactADRESSE POSTALE
IMGT®, the international ImMunoGeneTics information system®,
Laboratoire d'ImmunoGénétique Moléculaire (LIGM),
Institut de Génétique Humaine (IGH), UMR 9002 CNRS-Université de Montpellier,
141 rue de la Cardonille, cedex 5
34396 Montpellier cedex 5, France
WEB SITE http://www.imgt.org

Responsables
DIRECTEUR EXECUTIF EMERITE
Marie-Paule Lefranc
DIRECTEUR
Sofia Kossida
RESPONSABLE INFORMATIQUE
Patrice Duroux
RESPONSABLE BIOINFORMATIQUE
Véronique Giudicelli
Divers
POLE REGIONAL
IFB Grand Sud
CERTIFICATIONS
ISO 9001
NFX 50-900
IQuaRe
HON Certification




LABELLISATION
Internationale:
- Membre Académique Institutionnel de l'International Medical Informatics Association (IMIA).
- Membre de la Global Alliance for the Genomics and Health (GA4GH) depuis 2015.
- RIO depuis sa création en 2001, GIS IBiSA depuis sa création en 2007, ReNaBi depuis sa création.
- Labex MabImprove [ANR-10-LABX-53-01], GDR CNRS n° 3003 Molecular Bioinformatics (BiM), GDR CNRS n° 3260 Antibodies and Therapeutic targeting (ACCITH).
- Plateforme Bioinformatique du Cancéropôle Grand Sud-Ouest (GSO).
- BioCampus, Unité Mixte de Service (UMS 3426 CNRS - US 09 INSERM - UM), Montpellier.
- Grand Plateau Technique pour la Recherche et l'Innovation Languedoc-Roussillon (GPTR), depuis la création des GPTR en 2005.
IMGT® est une marque enregistrée CNRS (registered trademark) (Union Européenne, Canada, Etats-Unis).
STRUCTURE
CNRS
University of Montpellier


Equipe
Kossida Sofia, PU, IMGT director since 2015, University of Montpellier
Lefranc Marie-Paule, IMGT Founder and director (1989-2014), Professor Emeritus University of Montpellier
(2015-2019)
Albani Alexandre, IE CDD CNRS
Bertignac Morgane, IE CDD CNRS
Cherouali Karima, IE CDD CNRS
Duroux Patrice, IR CNRS
Elazami Elhassani Merouane, PhD student
Folch Géraldine, IE CNRS
Giudicelli Véronique, IR University of Montpellier
Goret Agathe, IE CDD CNRS
Jabado-michaloud Joumana
Kushwaha Anjana, IE CDD CNRS
Nguefack ngoune Viviane, IE CDD CNRS
Pegorier Perrine, IE CDD CNRS
Rollin Mael, IE CDD CNRS
Sanou Gaoussou, IE CDD CNRS
Tran Anna, PhD student
Viart Benjamin, IR CDD CNRS
Infrastructure
DESCRIPTION DES SERVEURS- Serveur DELL/PE6850 (lambda), 2 processeurs, 4 coeurs, 8 Go RAM, bases de données (IMGT, localisation IGH), 2006
- Serveur DELL/PE6850 (gamma), 4 processeurs, 16 coeurs, 32 Go RAM, calcul (IMGT, localisation IGH), 2007
- Serveur DELL/R710 (kappa2), 2 processeurs, 12 coeurs, 32 Go RAM, utilisateurs (IMGT, localisation IGH), 2010
- Serveur DELL/R510 (delta), 1 processeur, 6 coeurs, 1 Go RAM, partage de fichiers (IMGT, localisation IGH), 2012
- Serveur DELL/R420 (omega), 1 processeur, 6 coeurs, 64 Go RAM, web (IMGT, localisation IGH), 2014
- Serveur DELL/R530 (KOS-2015S01), 1 processeur, 6 coeurs, 32 Go RAM, sauvegardes (IMGT, localisation DR13), 2015
- Serveur DELL/R630 (KOS-2015S02), 1 processeur, 16 coeurs, 96 Go RAM, calcul (IMGT, localisation DR13), 2015
- 8 machines virtuelles (lef-beta01 à lef-beta08) web, calcul.
En réponse aux appels à projets du GENCI (Grand Equipement National de Calcul Intensif), les
heures de calcul suivantes ont été attribuées à IMGT® sur le Centre de Calcul
du CINES [allocation 036029]:
2009 : 60.000h (IBM)
2010 : 50.000h (IBM) et 80.000 (SGI) - Allocation c2010036029
2011 : 72.000h (IBM) et 80.000h (SGI) - Allocation c2011036029
2012 : 50.000h (IBM) et 50.000h (SGI) - Allocation c2012036029
2013 : 45.000h (IBM) et 320.000h (SGI) - Allocation c2013036029
2014 : 70.000h (IBM) et 304.000h (SGI) - Allocation c2014036029
2015 : 300.000 (TGCC BULL), 10.000 (CINES IBM) et 500.000 (CINES BULL) - Allocation x2015036029
2016 : 200.000 (TGCC BULL) et 275.000 (CINES BULL) - Allocation x2016036029
2017 : 150.000 (TGCC BULL) et 350.000 (CINES BULL) - Allocation A0010306029
2018 : 300.000 (TGCC BULL) et 500.000 (CINES BULL) - Allocation A0030306029
2019 : 25.000 (TGCC BULL) et 800.000 (CINES BULL) - Allocation A0050306029
2020 : 50.000 (TGCC BULL) et 350.000 (CINES BULL) - Allocation A0070306029
IMGT a également accès au HPC@LR.
Données
BASES DE DONNEES
IMGT® comprend sept bases de données:
1. IMGT/LIGM-DB est la base de données internationale pour les séquences nucléotidiques
d'IG et TR de l'homme et des autres espèces de vertébrés avec traduction des
séquences entièrement annotées.
2. IMGT/PRIMER-DB.
3. IMGT/GENE-DB. Des liens réciproques existent entre IMGT/GENE-DB et la base de données
'Gene' au NCBI (ceci est le seul exemple de liens directs effectués par Gene au NCBI sur une base
de données externe).
4. IMGT/3Dstructure-DB.
5. IMGT/2Dstructure-DB.
6. IMGT/CLL-DB, la base de données IMGT® des séquences IG de patients atteints de
leucémie lymphoïde chronique, créée en 2007.
7. IMGT/mAb-DB, la base de données IMGT® des anticorps monoclonaux et des protéines de
fusion à visée thérapeutique, créée en 2009.
RESSOURCES WEB
Les ressources Web IMGT® comprennent plus de 20.000 pages, organisées en plusieurs
sections:
1. IMGT Scientific chart fournit le vocabulaire contrôlé et les règles d'annotation
basés sur IMGT-ONTOLOGY.
2. IMGT Repertoire fournit une interface conviviale aux données soigneusement expertisées qui
concernent le génome, le protéome, le polymorphisme et la structure des IG et TR de l'homme et
des autres vertébrés.
3. IMGT Index, IMGT Education, IMGT Aide-mémoire, IMGT Bloc-notes.
4. IMGT Immunoinformatics page.
5. IMGT Medical page fournit aux cliniciens les informations d'IMGT® sur les bases, outils et
données d'intérêt médical.
6. IMGT Veterinary page fournit aux vétérinaires les informations d'IMGT® sur les
génomes, séquences et structures 3D des IG et TR des espèces animales.
7. IMGT Biotechnology page fournit un ensemble d'informations sur l'ingénierie des anticorps (phage
display), les anticorps de camélidés, l'humanisation des anticorps monoclonaux
thérapeutiques.
Outils
IMGT® OUTILS EN LIGNEIMGT® comprend dix-sept outils interactifs en ligne pour l'analyse des séquences (5), des gènes (7), des domaines protéiques (3) et des données structurales (2).
ANALYSE DES SEQUENCES
1. IMGT/V-QUEST, l'outil le plus utilisé d'IMGT®, intègre
2. IMGT/JunctionAnalysis.
3. IMGT/HighV-QUEST, pour les séquences IG et TR issues du Next Generation Sequencing (NGS).
4. IMGT/Allele-Align permet la comparaison de deux allèles.
5. IMGT/PhyloGene calcule et représente les arbres phylogénétiques des séquences
nucléotidiques d'IG et de TR.
ANALYSE DES GENES
1. IMGT/GeneView,
2. IMGT/GeneSearch,
3. IMGT/LocusView
4. IMGT/CloneSearch
5. IMGT/GeneInfo
6. IMGT/GeneFrequency
7. IMGT/LIGMotif.
ANALYSE DES DOMAINES PROTEIQUES
1. IMGT/DomainDisplay affiche les domaines protéiques selon la numérotation unique IMGT.
2. IMGT/DomainGapAlign permet de créer les « gaps » selon la numérotation unique
IMGT.
3. IMGT/Collier-de-Perles fournit la représentation standardisée « IMGT Colliers de
Perles » des domaines.
ANALYSE DES DONNEES STRUCTURALES
1. IMGT/StructuralQuery permet de faire des requêtes basées sur une description
structurale.
2. IMGT/DomainSuperimpose superpose deux structures 3D de domaines.
Expertise
Domaines d'activité
IMMUNOGENETIQUE
IMMUNOINFORMATIQUE
IMMUNOLOGIE
ANTICORPS THERAPEUTIQUES
BIOMEDICAL
BIOTECHNOLOGIE
DIAGNOSTIC
Description des expertises
IMGT®, the international ImMunoGeneTics information system® http://www.imgt.org, est la référence internationale en immunogénétique et immunoinformatique. IMGT® est une marque déposée du CNRS (Union Européenne, Canada et Etats-Unis). IMGT® est membre institutionnel académique de l'International Medical Informatics Association (IMIA) depuis 2006. IMGT® est spécialisé dans les séquences, structures et données génétiques des immunoglobulines (IG), des récepteurs T (TR), des protéines majeures d'histocompatibilité (MH) des vertébrés, des protéines apparentées du système immunitaire (RPI, pour Related Proteins of the Immune system) des superfamilles IgSF et MhSF des vertébrés et invertébrés, des protéines de fusion pour applications immunologiques (FPIA, pour fusion proteins for immune applications)et des protéines composites pour applications cliniques (CPCA, pour composite proteins for clinical applications). IMGT® est composé de sept bases de données (séquences, gènes et structures tridimensionnelles (3D)), de dix-sept outils interactifs et de plus de 15.000 pages de ressources Web .
IMGT® est reconnu internationalement pour la richesse et la qualité de ses données scientifiques et son interface conviviale. Le nombre d'équipes utilisatrices d'IMGT® est supérieur à 80.000. Ces équipes académiques et industrielles appartiennent à des domaines d'activité de recherche professionnelle variés et sont réparties dans le monde entier, à parts égales entre les Etats-Unis et Canada, l'Europe et le reste du monde. Ceci représente plus de 200.000 requêtes par mois et un nombre de sessions de travail sans cesse croissant (300.000 en 2015).
IMGT® est utilisé par des chercheurs d'équipes académiques et industrielles dans de
multiples domaines de recherche:
(1) recherche fondamentale,
(2) recherche médicale (analyse des répertoires des anticorps et des sites de reconnaissance
des récepteurs T dans les réponses immunitaires normales (infections, cancers), les
réponses anormales (maladies autoimmunes, immunodéficiences) et dans les syndromes
prolifératifs (leucémies, lymphomes, myélomes)),
(3) recherche vétérinaire (étude des répertoires des IG et TR dans les espèces
domestiques et sauvages),
(4) recherche génomique (étude de la diversité et de l'évolution des gènes de
la réponse immunitaire adaptative),
(5) recherche en biologie structurale (évolution des domaines des protéines des superfamilles
IgSF et MhcSF des vertébrés et invertébrés),
(6) biotechnologies relatives aux projets de l'Human Proteome Organisation (HUPO) et à
l'ingénierie des anticorps (single chain Fragment variable scFv, Fab, banques combinatoires, phage
display, anticorps chimériques, humanisés et humains),
(7) recherche pour le diagnostic, le pronostic et le suivi thérapeutique des leucémies,
lymphomes et myélomes (identification du ou des clone(s) malin(s) et évaluation de la maladie
résiduelle), (8) approches thérapeutiques (greffes, immunothérapie, vaccinologie).
Formations
FORMATION PROFESSIONNELLE
1. Atelier technologique. Biologie moléculaire appliquée aux anticorps: Analyse des
séquences et structures d'anticorps à l'aide des outils IMGT®
Montpellier 4-5 juin 2012
Responsable Scientifique: Marie-Paule Lefranc
Intervenants: Marie-Paule Lefranc, Véronique Giudicelli
Pré-requis: Biologistes (biologie moléculaire et immunologie) ayant un projet d'analyse de
séquences d'anticorps
Public: Etudiants, techniciens, ingénieurs et chercheurs
Nombre de participants: 12
Programme Formation théorique :
- Analyse du gène à la protéine
- Synthèse des immunoglobulines
- Concepts d'identification, de description, de classification et de numérotation
d'IMGT-ONTOLOGY
Programme Formation pratique : Utilisation en ligne des outils IMGT® :
- IMGT/V-QUEST
- IMGT/JunctionAnalysis
- IMGT/Collier-de-Perles
- IMGT/DomainGapAlign
- IMGT/3Dstructure-DB
2. Atelier technologique de BioCampus Montpellier. Utilisation des bases de données et outils
IMGT® pour l'ingénierie et l'humanisation des anticorps
Montpellier, 6-7 juin 2013
Responsable Scientifique: Marie-Paule Lefranc
Intervenants: Marie-Paule Lefranc, Souphatta Sasorith
3. Atelier technologique de BioCampus Montpellier. Utilisation des bases de données et outils
IMGT® pour l'ingénierie et l'humanisation des anticorps
Montpellier, 5-6 juin 2014
Responsable Scientifique: Marie-Paule Lefranc
Intervenants: Marie-Paule Lefranc, Souphatta Sasorith
4. Atelier technologique de BioCampus Montpellier. Vue d'ensemble des ressources IMGT®
Montpellier, 4 juin 2019
Responsable Scientifique: Sofia Kossida
Intervenants: Sofia Kossida, Véronique Giudicelli
FORMATION UNIVERSITAIRE
Master de Biologie-Santé, Université de Montpellier:
- parcours BIOTIN, «Mangement de projet et Innovation en Biotechnologie», http://www.master-biotin.fr/
- parcours Biophysique, Structures et Systèmes, http://www.masterbs.univ-montp2.fr/
- Parcours «Bioinformatique, Connaissances, Données», http://www2.lirmm.fr/bcd/
- Parcours Sciences et procédés des agroressources pour l'alimentation et l'environnement (SPA2E)
Module "Bioinformatique et Ontologies" (annuel de 2007 à 2014)
Responsable: Marie-Paule Lefranc
Maison des Ecoles Doctorales de Montpellier
Immunoinformatics - Bioinformatics of antibodies
University of Science and Technology of Hanoi.
Faculty of Biotechnology - Pharmacology, Hanoi, Vietnam
Consortium USTH (participation of the University of Montpellier)
(in English, BP41, Master 2, 40h, 4 ECTS, students from Biomedicine and from Drug development)
Prof. Marie-Paule Lefranc
Teaching Unit Content
1. Knowledge of databases for sequences and structures of antibodies.
2. Using tools for sequence analysis: applications to the study of repertoires of antibody recognition
sites and to the study of specificities (autoimmune diseases, infectious diseases, AIDS, leukemias,
lymphomas, myelomas...).
3. Bioinformatic structural analysis for the humanization of monoclonal antibodies for therapeutic
purposes (IMGT Collier de Perles methodology).
4. Structural analysis of antibody fragments and their recognition sites for the discovery of new
drugs.
This teaching unit is based in particular on the tools and databases developed by IMGT®, the
international ImMunoGeneTics information system ®, http://www.imgt.org, the international reference
system used by many pharmaceutical companies (JANSSEN Centocor Research and Development Inc. Johnson
& Johnson USA, AMGEN Inc. USA, Sanofi-Aventis GmbH Germany, Merck & Co. Inc. USA, Chugai
Pharmaceutical Co. Ltd. Japan, Astellas Pharma Inc. Japan, Agensys Inc. USA, Merck Serono SA
Switzerland, F. HOFFMAN-LA ROCHE Switzerland, etc..).
The teaching requires a PC connected to Internet per student.
Ouverture
Description de la répartitionACCES A IMGT®
Accès en ligne des 7 bases de données, 17 outils et aux 15.000 pages de ressources Web.
Caractère international de l'accès.
L'accès à IMGT® est libre, gratuit pour les académiques, sous licences et contrats avec le CNRS pour les industriels.
1. Site Web (http://www.imgt.org).
2. Distribution des données d'IMGT/LIGM-DB sur les serveurs IMGT
(http://www.imgt.org/download/LIGM-DB) et par FTP sur les serveurs de l'EBI
(ftp://ftp.ebi.ac.uk/pub/databases/imgt/LIGM-DB).
3. Distribution des données d'IMGT/GENE-DB sur les serveurs IMGT
(http://www.imgt.org/download/GENE-DB/).
4. Distribution des données d'IMGT/3Dstructure-DB sur les serveurs
IMGT(http://www.imgt.org/download/3Dstructure-DB/).
5. Accès aux données d'IMGT/LIGM-DB
- par SRS sur le serveur du Deutsche Krebsforschungszentrum (DKFZ) Heidelberg, Allemagne,
- par ARSA au DNA Data Bank of Japan (DDBJ).
6. Accès aux données par DAS et Dbfetch à l'EBI et LinkOut au NCBI.
7. Comparaison de séquences par rapport aux séquences de références d'IMGT/GENE-DB,
d'IMGT/DomainDisplay, d'IMGT/3Dstructure-DB et des séquences de domaines par BLAST sur le serveur
IMGT (IMGT/BlastSearch Query page).
8. Comparaison de séquences par BLAST et FASTA, par rapport à celles de IMGT/LIGM-DB, sur les
serveurs de l'EBI et de l'Institut Pasteur.
9. Références croisées avec HGNC, NCBI Gene, Vega, UniProt, Sequence Ontology.
10. Aides spécifiques et conseils aux utilisateurs et API.
Répartition des utilisateurs
INTERNATIONAUX 99 %
NATIONAUX 1 %
Publications
PUBLICATIONS EXTERNES2015
Aouinti S, Malouche D, Giudicelli V, Kossida S, Lefranc M-P.
IMGT/HighV-QUEST statistical significance of IMGT clonotype (AA) diversity per gene for standardized
comparisons of next generation sequencing immunoprofiles of immunoglobulins and T cell receptors.
PLoS ONE. 2015 Nov 5;10(11):e0142353. doi: 10.1371/journal.pone.0142353. eCollection 2015. Free Article PMID: 26540440 Correction: PLoS ONE 2016 Jan
5;11(1): e0146702. doi: 10.1371/journal.pone.0146702 View correction.
PMID: 26731095.
Piccinni B, Massari S, Caputi Jambrenghi A, Giannico F, Lefranc M-P, Ciccarese S, Antonacci R.
Sheep (Ovis aries) T cell receptor alpha (TRA) and delta (TRD) genes and genomic organization of the
TRA/TRD locus.
BMC Genomics. 2015 Sep 18;16(1):709. doi: 10.1186/s12864-015-1790-z. PMID: 26383271
Giudicelli V, Duroux P, Lavoie A, Aouinti S, Lefranc M-P, Kossida S.
From IMGT-ONTOLOGY to IMGT/HighV-QUEST for NGS immunoglobulin (IG) and T cell receptor (TR) repertoires
in autoimmune and infectious diseases
Autoimmun Infec Dis. 2015 Aug 10. 1(1): doi: 10.16966/aidoa.103. Free
Article.
Lefranc M-P.
Antibody Informatics: IMGT, the International ImMunoGeneTics Information System, In: Crowe J, Boraschi
D, Rappuoli R (Eds)
Antibodies for Infectious Diseases. ASM Press, Washington, DC. doi: 10.1128/microbiolspec.
AID-0001-2012, 2015, pp. 363-379.
Li L, Wang XH, Williams C, Volsky B, Steczko O, Seaman MS, Luthra K, Nyambi P, Nadas A, Giudicelli V,
Lefranc M-P, Zolla-Pazner S, Gorny MK.
A broad range of mutations in HIV-1 neutralizing human monoclonal antibodies specific for V2, V3, and
the CD4 binding site.
Mol Immunol. 2015 May 18;66(2):364-374. doi: 10.1016/j.molimm.2015.04.011. PMID: 25965315
Baliakas P, Agathangelidis A, Hadzidimitriou A, Sutton LA, Minga E, Tsanousa A, Scarfò L, Davis Z,
Yan XJ, Shanafelt T, Plevova K, Sandberg Y, Vojdeman FJ, Boudjogra M, Tzenou T, Chatzouli M, Chu CC,
Veronese S, Gardiner A, Mansouri L, Smedby KE, Pedersen LB, Moreno D, Van Lom K, Giudicelli V, Francova
HS, Nguyen-Khac F, Panagiotidis P, Juliusson G, Angelis L, Anagnostopoulos A, Lefranc M-P, Facco M,
Trentin L, Catherwood M, Montillo M, Geisler CH, Langerak AW, Pospisilova S, Chiorazzi N, Oscier D,
Jelinek DF, Darzentas N, Belessi C, Davi F, Ghia P, Rosenquist R, Stamatopoulos K.
Not all IGHV3-21 chronic lymphocytic leukemias are equal: prognostic considerations.
Blood. 2015 Jan 29;125(5):856-9. doi: 10.1182/blood-2014-09-600874. PMID: 25634617
Xochelli A, Agathangelidis A, Kavakiotis I, Minga E, Sutton LA, Baliakas P, Chouvarda I, Giudicelli V,
Vlahavas I, Maglaveras N, Bonello L, Trentin L, Tedeschi A, Panagiotidis P, Geisler C, Langerak AW,
Pospisilova S, Jelinek DF, Oscier D, Chiorazzi N, Darzentas N, Davi F, Ghia P, Rosenquist R,
Hadzidimitriou A, Belessi C, Lefranc M-P, Stamatopoulos K.
Immunoglobulin heavy variable (IGHV) genes and alleles: new entities, new names and implications
for research and prognostication in chronic lymphocytic leukemia.
Immunogenetics. 2015 67(1):61-6.doi: 10.1007/s00251-014-0812-3. PMID: 25388851
Lefranc M-P, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H,
Houles A, Paysan-Lafosse T, Hadi-Saljoqi S, Sasorith S, Lefranc G, Kossida S.
IMGT®, the international ImMunoGeneTics information system® 25 years on.
Nucleic Acids Res. 2015 Jan;43(Database issue):D413-22. doi: 10.1093/nar/gku1056. Free Article.
PMID: 25378316
2014
Lefranc M-P.
Immunoglobulins: 25 years of Immunoinformatics and IMGT-ONTOLOGY.
Biomolecules. 2014, 4(4), 1102-1139; doi:10.3390/biom4041102 Open access. PMID: 25521638
Lefranc M-P.
IMGT® immunoglobulin repertoire analysis and antibody humanization.
In: Alt, F.W, Honjo, T, Radbruch A. and Reth, M. (Eds.), Molecular Biology of B cells, Second
edition, Academic Press, Elsevier Ltd, London, UK, Chapter 26, 2014, pp. 481-514. dx.doi.org, ISBN :
978-0-12-397933-9.
Lefranc M-P.
How to use IMGT® for therapeutic antibody engineering
In: Dübel S, Reichert J (Eds), Handbook of Therapeutic Antibodies (4 vol.), Second edition,
Wiley, Volume 1: Defining the right antibody composition, Chapter 10, 2014, pp. 229-264.
Shirai H, Prades C, Vita R, Marcatili P, Popovic B, Xu J, Overington JP, Hirayama K, Soga S, Tsunoyama K,
Clark D, Lefranc M-P, Ikeda K.
Antibody informatics for drug discovery.
Biochim Biophys Acta. 2014 Nov;1844(11):2002-2015. pii: S1570-9639(14)00174-5. doi:
10.1016/j.bbapap.2014.07.006. PMID: 25110827
Lefranc M-P.
Immunoinformatics of the V, C, and G Domains: IMGT® Definitive System for IG, TR and IgSF,
MH, and MhSF
Methods Mol Biol. 2014;1184:59-107. doi: 10.1007/978-1-4939-1115-8_4. PMID: 25048119
Gogoladze G, Grigolava M, Vishnepolsky B, Chubinidze M, Duroux P, Lefranc M-P, Pirtskhalava M.
DBAASP: Database of Antimicrobial Activity and Structure of Peptides
FEMS Microbiol Lett. 2014 Aug; 357(1):63-68. doi: 10.1111/1574-6968.12489. PMID: 24888447
Ciccarese S, Vaccarelli G, Lefranc M-P, Tasco G, Consiglio A, Casadio R, Linguiti G, Antonacci R.
Characteristics of the somatic hypermutation in the Camelus dromedarius T cell receptor gamma
(TRG) and delta (TRD) variable domains.
Dev Comp Immunol. 2014 May Oct;46(2):300-313. doi: 10.1016/j.dci.2014.05.001. PMID: 24836674
Alamyar A, Giudicelli V, Duroux P, Lefranc M-P.
Antibody V and C domain sequence, structure and interaction analysis with special reference to
IMGT®
In: Ossipow V and Fischer N. (Eds.), Monoclonal antibodies: Methods and Protocols, Second edition.
Humana Press, Springer Science+Business Media, LLC, New York, USA. Methods Mol Biol. 2014.1131:337-81.
doi: 10.1007/978-1-62703-992-5_21. PMID:
24515476
Lefranc M-P.
Antibody informatics: IMGT®, the international ImMunoGeneTics information system®, the
international ImMunoGeneTics information system®
Microbiol Spectrum 2014, 2(2):AID-0001-2012. doi:10.1128/microbiolspec.AID-0001-2012.
Lefranc M-P.
Immunoglobulin (IG) and T cell receptor genes (TR): IMGT® and the birth and rise of
immunoinformatics.
Front Immunol. 2014 Feb 05;5:22. doi: 10.3389/fimmu.2014.00022. Open access. PMID: 24600447
Ouled-Haddou H, Ghamlouch H, Regnier A, Trudel S, Herent D, Lefranc M-P, Marolleau J, Gubler B.
Characterization of a new V gene replacement in the absence of activation-induced cytidine
deaminase and its contribution to human BcR diversity.
Immunology. 2014 Feb;141(2):268-75. doi: 10.1111/imm.12192. PMID: 24134819
2013
Six A, Mariotti-Ferrandiz ME, Chaara W, Magadan S, Pham H-P, Lefranc M-P, Mora T, Thomas-Vaslin V,
Walczak AM, Boudinot P.
The past, present and future of immune repertoire biology - the rise of next-generation repertoire
analysis.
Front Immunol. 2013 Nov 27;4:413. doi: 10.3389/fimmu.2013.00413 Open access. PMID: 24348479
Lefranc M-P.
IMGT Collier de Perles
In: Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H (Eds), Encyclopedia of Systems Biology, New York:
Springer Science+Business Media, LLC, 2013, pp. 944-952.
Lefranc M-P.
IMGT unique numbering
In: Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H (Eds), Encyclopedia of Systems Biology, New York:
Springer Science+Business Media, LLC, 2013, pp. 952-959.
Giudicelli V, Lefranc M-P.
IMGT-ONTOLOGY
In: Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H (Eds), Encyclopedia of Systems Biology, New York:
Springer Science+Business Media, LLC, 2013, pp. 964-972.
Lefranc M-P.
IMGT® information system
In: Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H (Eds), Encyclopedia of Systems Biology, Springer
Science+Business Media, LLC, New York, 2013, pp. 959-964.
Li S, Lefranc M-P, Miles JJ, Alamyar E, Giudicelli V, Duroux, P, Freeman JD, Corbin VDA, Scheerlinck J-P,
Frohman MA, Cameron PU, Plebanski M, Loveland B, Burrows SR, Papenfuss AT, Gowans EJ.
IMGT/HighV-QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation
repertoire immunoprofiling
Nat. Commun. 4:2333 doi: 10.1038/ncomms3333 (2013) Open access. PMID: 23995877
Lefranc M-P, Lefranc G.
Consanguinity
In: Stanley Maloy and Kelly Hughes, editors. Brenner's Encyclopedia of Genetics 2nd edition, Vol 2. San
Diego: Academic Press; 2013. pp. 158-162.
Castro R, Jouneau L, Pham HP, Bouchez O, Giudicelli V, Lefranc M-P, Quillet E, Benmansour A, Cazals F,
Six A, Fillatreau S, Sunyer O, Boudinot P.
Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection
PLoS Pathog. 2013 Jan;9(1):e1003098. doi: 10.1371/journal.ppat.1003098. PMID: 23326228
Created 31/03/2016 (G. Folch)
IMGT server environment 
Computing environment
The IMGT® information system is localized at the Laboratoire
d'ImmunoGénétique Moléculaire (LIGM), Institut de Génétique
Humaine (IGH) of the
Centre National de la Recherche Scientifique (CNRS), Montpellier, France.
IMGT/HighV-QUEST HPC resources are hosted at the Centre Informatique
National de l'Enseignement Supérieur (CINES).
Software environment
- The operating system is Linux.
- The Web servers are HTTP and Tomcat from The Apache Software Foundation.
- The databases are hosted on Sybase system (Sybase Inc.) and MySQL™.
- The programs are written in Java, Perl and Bourne shell.
Network environment
The Internet access is provided by the french academic network RENATER.
IMGT history 
The project of an integrated system in Immunogenetics has been started in 1989 by Marie-Paule Lefranc, Professor Université Montpellier 2, CNRS, Montpellier, France.
A prototype of the ImMunoGeneTics information system was developed in France for Immunoglobulin and T cell receptor sequences (LIGM-DB) by the Laboratoire d'ImmunoGénétique Moléculaire (LIGM, CNRS, Université Montpellier 2, Montpellier), in collaboration with the Centre National Universitaire Sud de Calcul (CNUSC, Montpellier, Sophie Creuzet and Denys Chaume) and the Laboratoire d'Informatique, de Robotique et de Micro-électronique de Montpellier (LIRMM, CNRS, Université Montpellier 2, Montpellier, Isabelle Mougenot and Patrice Déhais).
In 1992 the database became truly european with the collaboration of EMBL (Rainer Fuchs),
IFG (Werner Müller), ICRF (Julia Bodmer) and the obtention of a EU European Union BIOMED1
funding (BIOCT930038).
IMGT/LIGM-DB is available on the Web server of CINES at Montpellier since July 1995.
The first demonstration on-line was performed at the 9th International Congress of Immunology
at San Francisco, USA (23-29 July 1995).
IMGT/MHC-HLA, for human MH1 and MH2 (or HLA) sequences maintained by ANRI, London,
is on the EBI server since December 1998.
IMGT®, the international ImMunoGeneTics information system®, enhanced its development through the European Community 5th PCRDT funding QLG2-2000-01287 (collaboration between LIGM, EMBL-EBI, Cancer Research UK, EUROGENTEC and BPRC), and then ImmunoGrid of the EU 6th PCRDT (IST-2004-028069). Since 2008, IMGT® is part of ELIXIR.
IMGT significant dates
1989
- Creation of IMGT® at the Human Gene Mapping 10 Workshop (HGM10), New Haven (USA) (11-17 June 1989)
1993-1996
- European Union BIOMED1 funding (BIOCT930038)
1995
- IMGT/LIGM-DB on the Web, for the first time, at the 9th International Congress of Immunology (9,000 participants), San Francisco (USA) (23-29 July 1995)
1996-1999
- European Union BIOTECH2 funding (BIO4CT960037)
1997
- First IMGT Collier de Perles on the Web (April 1997)
- IMGT/V-QUEST on the Web (07 July 1997)
1998
- IMGT/MHC-HLA on the Web (December 1998)
1999
2000-2003
- European Union 5th PCRDT funding (QLG2-2000-01287)
- Nucleic Acids Res., 28:219-221 (2000), PMID: 10592230
2001
- IMGT® identified as operational RIO platform (Bioinformatics) at the French national level (CNRS, INSERM, CEA, INRA)
- IMGT/GeneSearch, IMGT/GeneView andIMGT/LocusView on the Web (17 April 2001)
- IMGT/JunctionAnalysis on the Web (17 june 2001)
- IMGT/3Dstructure-DB on the Web (01 October 2001)
- Nucleic Acids Res., 29:207-209 (2001), PMID: 11125093
2002
- IMGT/StructuralQuery on the Web (15 January 2002)
- IMGT/Phylogene on the Web (10 February 2002)
- IMGT/PRIMER-DB on the Web (22 February 2002)
- IMGT/MHC-NHP, IMGT/MHC-DLA and -FLA on the Web (April 2002)
- NetWatch of Science "Blueprints of Immunity" , Science, vol. 296, p. 1207 (17 May 2002)
- IMGT/AlleleAlign on the Web (3 June 2002)
2003
- Editorial "The International ImMunoGeneTics database IMGT", by Warr, G.W., Clem, W. and Söderhäll, K. Dev. Comp. Immunol. 27,1 (2003) (27 January 2003)
- IMGT/GENE-DB on the Web (31 January 2003)
- ADER Prize for IMGT/PRIMER-DB implementation
- Nucleic Acids Res., 31:307-310 (2003), PMID: 12520009
2004
- IMGT/GeneInfo on the Web (14 January 2003)
- IMGT 15 years ! at the 12th International Congress of Immunology, Montreal (Canada) (18-23 July 2004)
- IMGT-Choreography funded as BioSTIC project
- IMGT-ONTOLOGY for immunogenetics and immunoinformatics: In Silico Biol.
4:17-29 (2004). PMID: 15089751

- ACI IMPBio 2004 IMGT project
accepted

2005
- IMGT® on the Ensembl Genome Browser
- Guide Européen de l'innovation (in French).
- IMGT-Choreography for Immunogenetics and Immunoinformatics: In Silico
Biology, 5, 45-60 (2005). PMID: 15972004

- IMGT-Choreography figure
 , Nucleic Acids Res., 33:D593-597
(2005). PMID: 15608269
, Nucleic Acids Res., 33:D593-597
(2005). PMID: 15608269 - IMGT® NAR 2005 figure

2006
- BIOSYS, ImmunoGrid
- IMGT® 2007
2007
- IMGT® received the label IBiSA
- ACI IMPBio 2007 Poster

2008
- Formal IMGT-ONTOLOGY, IMGT-Kaleidoscope
2009
- IMGT/mAb-DB (4 December 2009)
- Nucleic Acids Res., 37:D1006-1012 (2009), PMID: 18978023
2010
- IMGT® is certified ISO 9001:2008 (15 October 2010)
- IMGT/HighV-QUEST on the Web (22 November 2010)
2014
- IMGT® is certified NFX 50-900 (8 October 2014)
2015
- Nucleic Acids Res., 43:D413-422 (2015), PMID: 25378316
2017
- IMGT® is certified ISO 9001:2015 and NFX 50-900:2016 (28 November 2017)
Demonstrations of IMGT
| 2011 | July 17-19 | 19th Annual International Conference on Intelligent Systems for Molecular Biology and 10th European Conference on Computational Biology | Vienna, Austria |
| 2010 | July 11-13 | 18th Annual International Conference on Intelligent Systems for Molecular Biology | Boston, USA |
| 2010 | May 18-21 | 14th Human Genome Meeting - Next Generation Genomics and Medicine | Montpellier, France |
| 2009 | June 27 - July 2 | 17th Annual International Conference on Intelligent Systems for Molecular Biology (ISMB) and 8th European Conference on Computational Biology | ECCBStockholm, Sweden |
| 2008 | September 22-26 | European Conference on Computational Biology | ECCBCagliari, Sardinia-Italy |
| 2004 | July 18-23 | 12th International Congress of Immunology and 4th Annual Conference of FOCIS | Montreal, Canada |
| 2002 | November 29 | Internet et Pédagogie Médicale, IPM | Lille, France |
| 2000 | October 16-22 | Fête de la Science | Montpellier, France |
| 1999 | October 21-23 | Science en Fête | Montpellier, France |
| 1998 | October 23 | Salon ETIC, Université Montpellier II | Montpellier, France |
| 1998 | January 4-9 | Pacific Symposium on Biocomputing | Hawaï, USA |
| 1997 | October 10-12 | Science en Fête | Montpellier, France |
| 1996 | October 11-13 | Science en Fête | Montpellier, France |
| 1995 | July 22-29 | 9th International Congress of Immunology | San Francisco, USA |
| 1994 | December 15-16 | Réunion d'Automne de la SFI | Montpellier, France |
| 1994 | June 14-17 | 12th European Immunology Meeting, EFIS 94 | Barcelona, Spain |
| 1993 | November 14-17 | Human Genome Mapping Workshop 93, HGM 93 | Kobe, Japan |
Reviews on IMGT
- IMGT®: un langage commun pour l'immunogénétique
- In: INNOVATION, l'Economie de la croissance, Septembre-Octobre 2007, n° 10, p. 80.
- IMGT®: un langage commun pour l'immunogénétique
- In: INNOVATION, l'Economie de la croissance, n° 6, Janvier-Février 2007, p. 37.
- Laboratoire d'ImmunoGénétique Moléculaire. IMGT®: un langage commun pour l'immunogénétique
- In: Guide Européen de l'Innovation, site Internet du magazine Référence Innovation.
- IMGT®: a common language for
immunogenetics
IMGT®: un langage commun pour l'immunogénétique - In: Reference INNOVATION, n° 5, November-December 2006, pp. 60-61.
- IMGT®: un langage commun pour l'immunogénétique
- In: Reference Business, Industries Sante/Agro, septembre/octobre 2006, n°11, p. 63.
- IMGT®: un langage commun pour l'immunogénétique
- In: Reference Business, Les industries de la santé, janvier/février 2006, n°8, p. 94.
- Cover of Tissue Antigens
- Tissue Antigens, Vol 65, 6 issues (January 2005 to July 2005).
- WebWatch BioTechniques "French collection"

- A WebWatch report on IMGT, by Kevin Ahern, BioTechniques Euro Edition, The Journal of Laboratory Technology for Bioresearch n° 66, p. 17 (July/August 2004).
- Portail Institut Pasteur. Médiathèque. La sélection de la semaine: The international ImMunoGeneTics information system : IMGT
- Novembre 2004.
- ADER Prize 2003 for IMGT/PRIMER-DB (LIGM and EUROGENTEC S.A.)
- Life Sciences Prize for "Innovation-Research-Enterprise" by the Association for Education and Research Development (ADER).
- The International ImMunoGeneTics database IMGT

- Editorial by Warr G.W., Clem W. and Söderhall K. in Developmental Comparative Immunology (DCI) for the launch of the IMGT Locus in Focus section in DCI, in January 2003. Dev Comp. Immunol. 27,1 (2003) PMID: 12477495.
- We thank Elsevier Science for allowing IMGT to make available, on the IMGT site, the PDF file of the Editorial.
- IMGT
- In: Nucleic Acids Research - The Molecular Biology Database Collection: 2003 update.
- NetWatch of Science "Blueprints of Immunity"

- A NetWatch report on IMGT, by Mitch Leslie, Science, vol. 296, n°5571, p.1207 (17 May 2002).
- Des nouvelles de l'IMGT, la base de données internationale en ImMunoGénéTique
- La Gazette du Laboratoire n°64, p. 21 (2002).
- Compiling immune system data, Infrastructures, Contract BI04-CT96-0037
- In: Biotechnology programme (1994-98) Project reports (Vol. 2), p. 84.
Quality of Life and management of living resources, European Commission EUR19405 (2001) ISBN: 92-894-0240-7. - Genome Biology web report on IMGT "A high-quality database of molecules of the immune system"
- A Genome Biology report by Christopher I. Thorpe.
- Report date: 1st September 1999 - Published: 17 March 2000.
Genome Biology 2000 1:reports202. - Cover page of Nucleic Acids Research For the 10th Anniversary of IMGT
- Nucl. Acids Res., vol. 27 n°1 (January 1999) Database issue.
- The cover features an example of IMGT/Colliers de Perles and 3D representation for T cell receptor V-REGIONs, available at IMGT, The international ImMunoGeneTics information system®, http://www.imgt.org, created in 1989 by Marie-Paule Lefranc, Montpellier, and described in the article on pp. 209-212 of this issue. Lefranc, M.-P. et al., Nucleic Acids Research, 27, 209-212 (1999) PMID: 9847182 LIGM:209.