IMGT Repertoire (IG and TR)
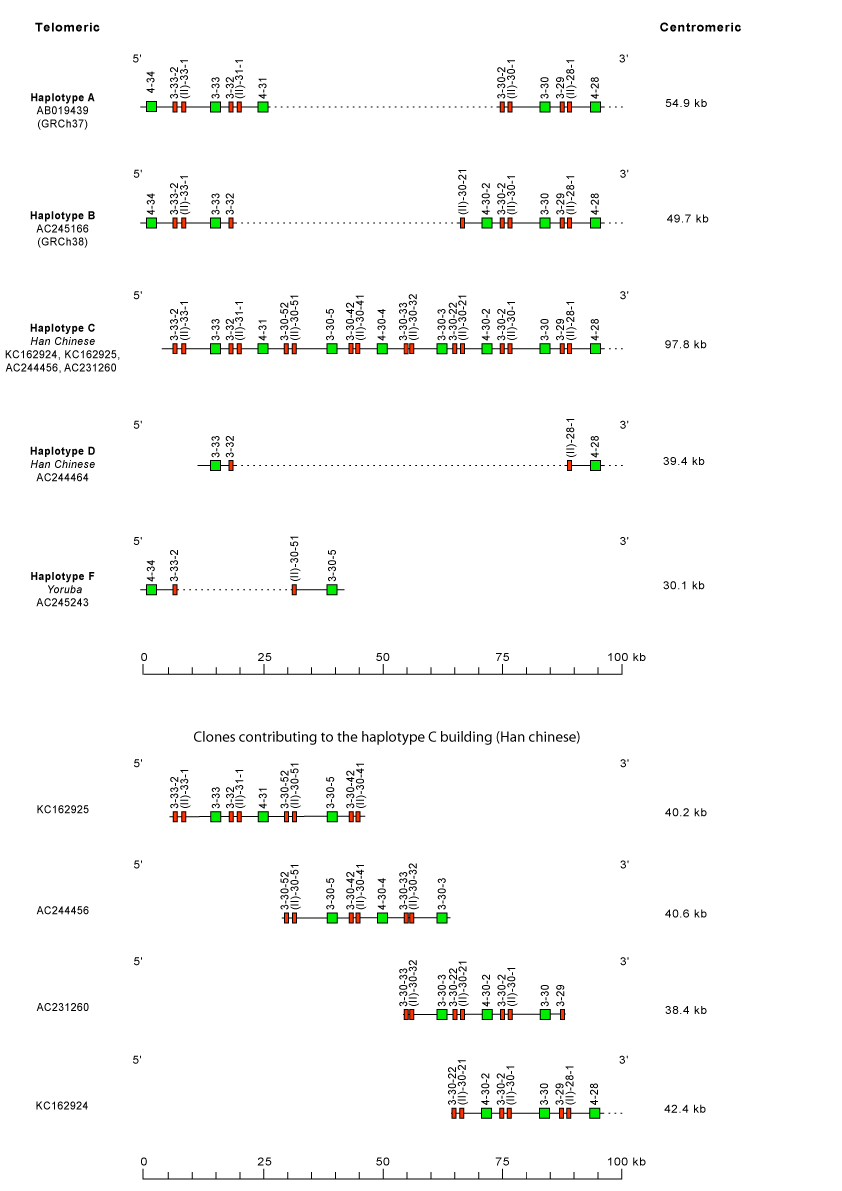
Locus representation: human (Homo sapiens) Polymorphism by insertion/deletion between IGHV4-34 and IGHV4-28 (haplotypes A to F) on chromosome 14 (14q32.33)
- Human (Homo sapiens) IGH locus on chromosome 14 (14q32.33)
- Human (Homo sapiens) IGH orphons on chromosome 15 (15q11.2)
- Human (Homo sapiens) IGH orphons on chromosome 16 (16p11.2)
'Haplotype' refers here to the description of gene copy number variation (CNV) polymorphism between IGHV4-34 and IGHV4-28.
Please note that it may correspond to an assembly of several clones of different origins as in 'haplotype' A, or to an incomplete description of the chromosomal region as in 'haplotypes' C, D, E and F.
Haplotype A is from GRCh37 and corresponds to the main line of IMGT Locus Representation [1].
Haplotype B is from GRCh38 and corresponds to BAC clone sequences [2] from the CHORI-17 BAC library (IMGT Lexique)
Distance between IGHV4-34 and IGHV4-28, lengths of sequences in regular line, are indicated for each haplotype in kilobases (kb).
Dotted lines indicate missing genes compared to the haplotype C.
Distance between IGHV3-33-2 and IGHV4-28, lengths of the individual clones (KC162925, AC244456, AC231260 and KC162924) which contributed to the building of the haplotype C (Han Chinese), are also indicated in kilobases (kb).
Human (Homo sapiens) Polymorphism by insertion/deletion between IGHV4-34 and IGHV4-28 (haplotypes A to F) on chromosome 14 (14q32.33)
Legend:
Colors are according to IMGT color menu for genes.
The boxes representing the genes are not to scale. Exons are not shown.
A double slash // indicates a gap in genome assembly. Distances in bp (base pair) or in kb (kilobase) associated with a double slash are taken
into account in the length of the lines and included in the numbers displayed at the right end of the lines.
Localization of functional genes are from [1]. Pseudogenes have been identified and annotated by E. Carillon (27/07/2015).
- [1] Lefranc, M.-P. and Lefranc, G. The Immunoglobulin FactsBook Academic Press, London, UK (458 pages), (2001). Print Book ISBN :9780124413511, eBook ISBN :9780080574479.
- [2] Watson C.T. et al., Am. J. Hum. Genet., 92, 530-546 (2013). PMID:23541343
- Created:
- 20/07/2015
- Last updated:
- 18/07/2024
- Authors:
- Imène Chentli, Emilie Carillon
