IMGT Repertoire (IG and TR)
Locus representation: western lowland gorilla (Gorilla gorilla gorilla) IGH locus on chromosome 14 Holistic IMGT reference
Western lowland gorilla (Gorilla gorilla gorilla) IGH locus on chromosome 14 Holistic IMGT reference
The orientation of the western lowland gorilla (Gorilla gorilla gorilla) IGH locus on chromosome 14 Holistic IMGT reference is reverse (REV).
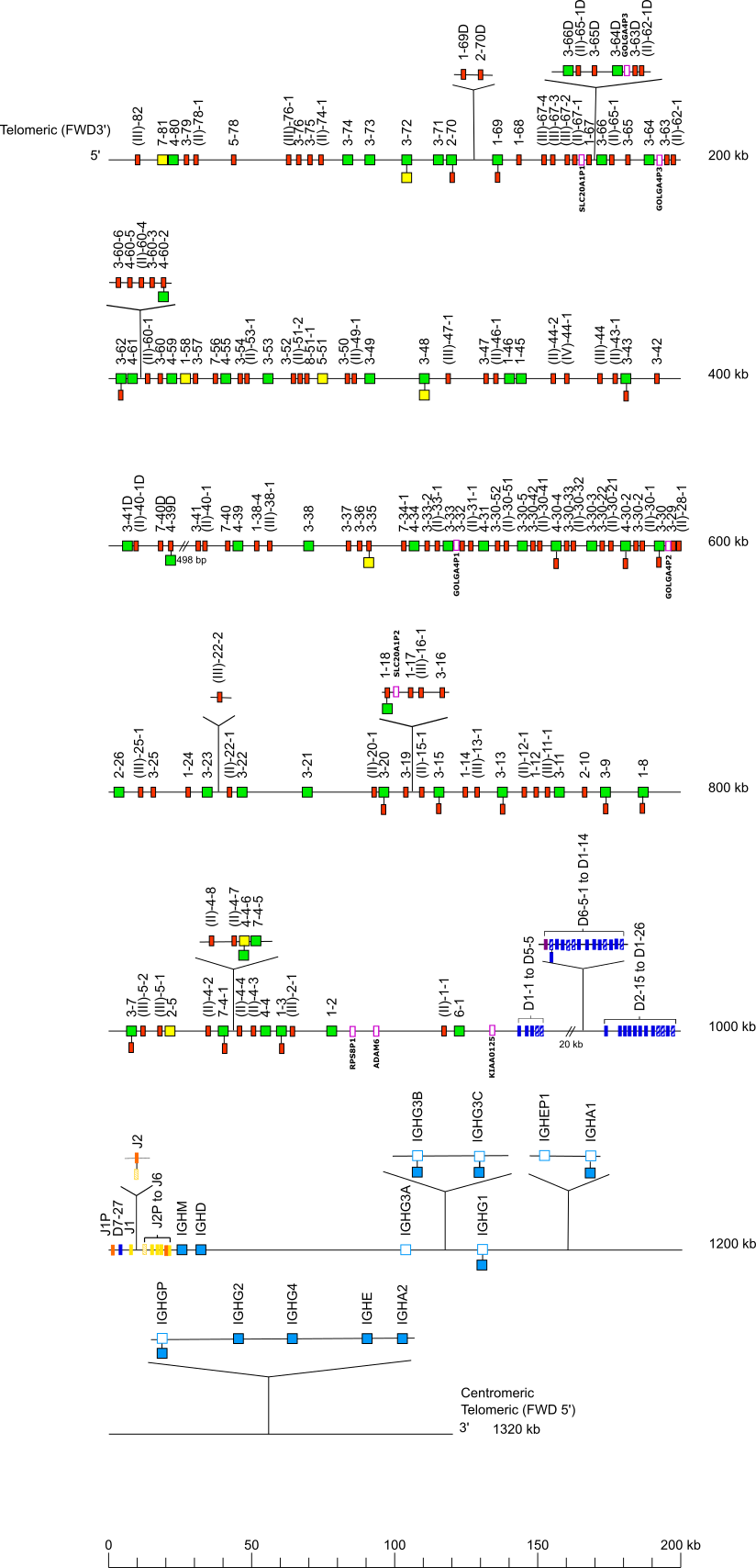
Legend:
Colors are according to IMGT color menu for genes.
The boxes representing the genes are not to scale. Exons are not shown.
A double slash // indicates a gap in genome assembly. Distances in bp (base pair) or in kb (kilobase) associated with a double slash are taken
into account in the length of the lines and included in the numbers displayed at the right end of the lines.
IGH gene names are according to the IMGT nomenclature [1].
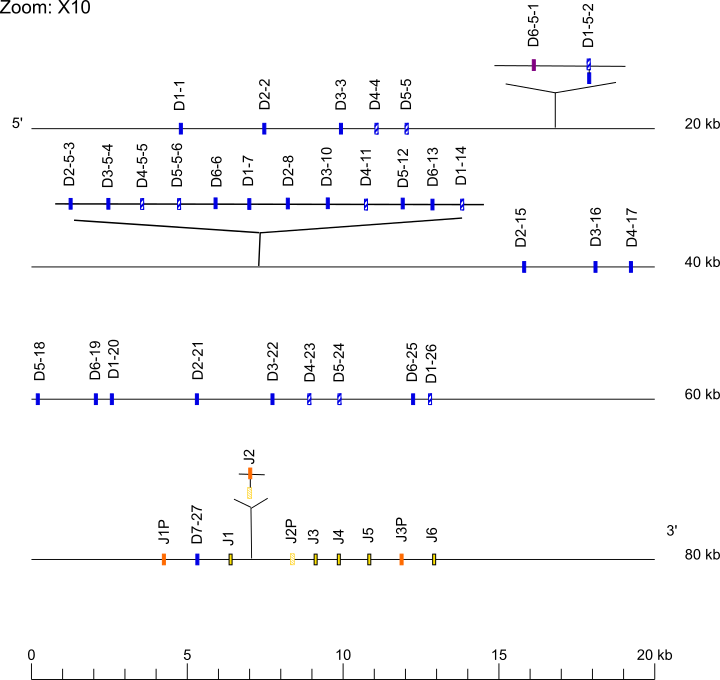
The IGHD6-5-1 to IGHD1-14 diversity genes are not found in the Kamilah_GGO_v0 assembly, there is a gap at this position (IMGT000074 accession number). However, the IGHD6-5-1 to IGHD1-14 are found in the Susie3 assembly (IMGT000078 accession number), in the NHGRI_mGorGor1-v1.1-0.2.freeze_mat assembly (IMGT000139 accession number) and in the NHGRI_mGorGor1-v1.1-0.2.freeze_pat assembly (IMGT000140 accession number).
The IGHV7-4-5, IGHV4-4-6, IGHV(II)-4-7, IGHV(II)-48, IGHV3-16, IGHV(III)-16-1, IGHV1-17, IGHV1-18, IGHV(III)-22-2, IGHV4-60-2, IGHV3-60-3, IGHV(II)-60-4, IGHV4-60-5, IGHV3-60-6, IGHV2-70D, IGHV1-69D, IGHD6-5-1, IGHD1-5-2, IGHD2-5-3, IGHD3-5-4, IGHD4-5-5, IGHD5-5-6, IGHD6-6, IGHD1-7, IGHD2-8, IGHD3-10, IGHD4-11, IGHD5-12, IGHD6-13, IGHD1-14, IGHJ2, IGHG3B, IGHG3C, IGHEP1, IGHA1 and IGHGP are found in the Susie3 assembly (IMGT000078 accession number).
The IGHV(II)-62-1D, IGHV3-63D, GOLGA4P3, IGHV3-64D, IGHV3-65D, IGHV(II)-65-1D and IGHV3-66D are found in the NHGRI_mGorGor1-v1.1-0.2.freeze_pat assembly (IMGT000140 accession number).
The IGHG3B, IGHG3C, IGHEP1, IGHA1, IGHGP, IGHG2, IGHG4, IGHE and IGHA2 are found in the Susie3 assembly (IMGT000078 accession number), in the NHGRI_mGorGor1-v1.1-0.2.freeze_mat assembly (IMGT000139 accession number) and in the NHGRI_mGorGor1-v1.1-0.2.freeze_pat assembly (IMGT000140 accession number).
Zoom for D and J genes
- [1] Lefranc M-P. Front Immunol. 2014 Feb 05;5:22. doi: 10.3389/fimmu.2014.00022. Open access.. PMID:24600447
- Gene table: Western lowland gorilla (Gorilla gorilla gorilla) IGHV
- Gene table: Western lowland gorilla (Gorilla gorilla gorilla) IGHD
- Gene table: Western lowland gorilla (Gorilla gorilla gorilla) IGHJ
- Gene table: Western lowland gorilla (Gorilla gorilla gorilla) IGHC
- Potential germline repertoire: Western lowland gorilla (Gorilla gorilla gorilla) IGHV, IGHD and IGHJ genes on chromosome 14
- Created:
- 15/12/2021
- Last updated:
- 11/07/2024
- Authors:
- Chahrazed Debbagh
