IMGT Repertoire (IG and TR)
Locus representation: House mouse (Mus musculus) IGH
House mouse (Mus musculus) IGH locus on chromosome 12 (12F2) strain C57BL/6
The orientation of the house mouse (Mus musculus) IGH locus on the chromosome 12 (12F2) strain C57BL/6 is reverse (REV).
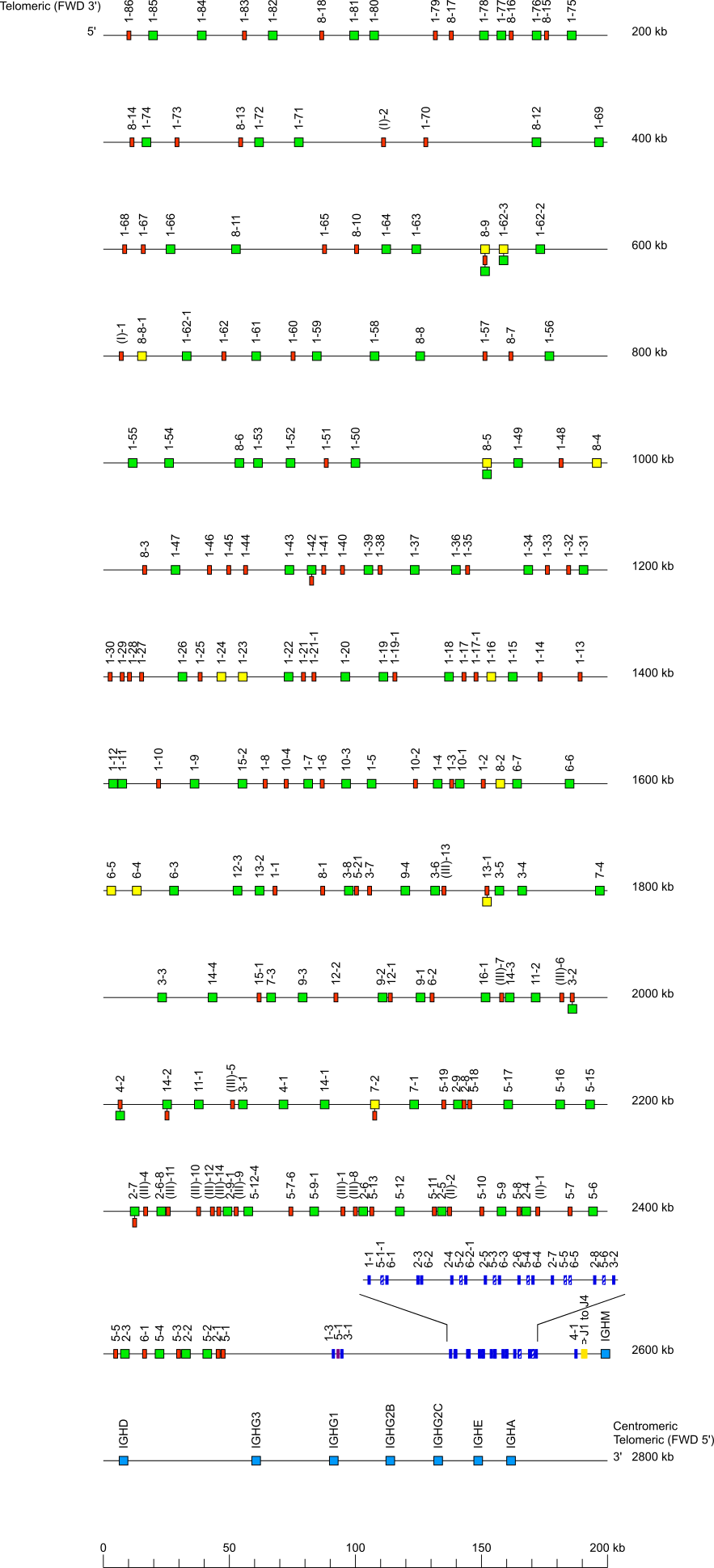
Legend:
Colors are according to IMGT color menu for genes.
The boxes representing the genes are not to scale. Exons are not shown.
A double slash // indicates a gap in genome assembly. Distances in bp (base pair) or in kb (kilobase) associated with a double slash are taken
into account in the length of the lines and included in the numbers displayed at the right end of the lines.
A dotted line ... indicates the distance in kb (kilobase) between the locus and its bornes (5' borne and the most 5' gene in the locus and/or 3'
borne and the most 3' gene in the locus) on the top and bottom lines, respectively. These distances are not represented at scale and/or are not
included in IMGT-LOCUS-UNIT and may not be included in the numbers displayed at the right ends of these two lines
.
SWITCH sequences are represented by a filled circle upstream of the IGHC genes.
The Mouse (Mus musculus) IGHV genes belong to 16 subgroups (Gene tables: Mouse IGHV) which are part of three clans (Figure IGHV clans). Clans comprise, respectively:
- clan I: IGHV1, IGHV9; IGHV14 and IGHV15 subgroup genes and one pseudogene IGHV(I)
- clan II: IGHV2, IGHV3, IGHV8 and IGHV12 subgroup genes and pseudogenes IGHV(II)
- clan III: IGHV4, IGHV5, IGHV6, IGHV7, IGHV10, IGHV11, IGHV13 and IGHV16 subgroup genes and pseudogenes IGHV(III).
Locus representation with only the functional genes: house mouse (Mus musculus) IGH

* When needed, indicate the presence of allele(s) with another functionality: see above house mouse (Mus musculus) IGH locus
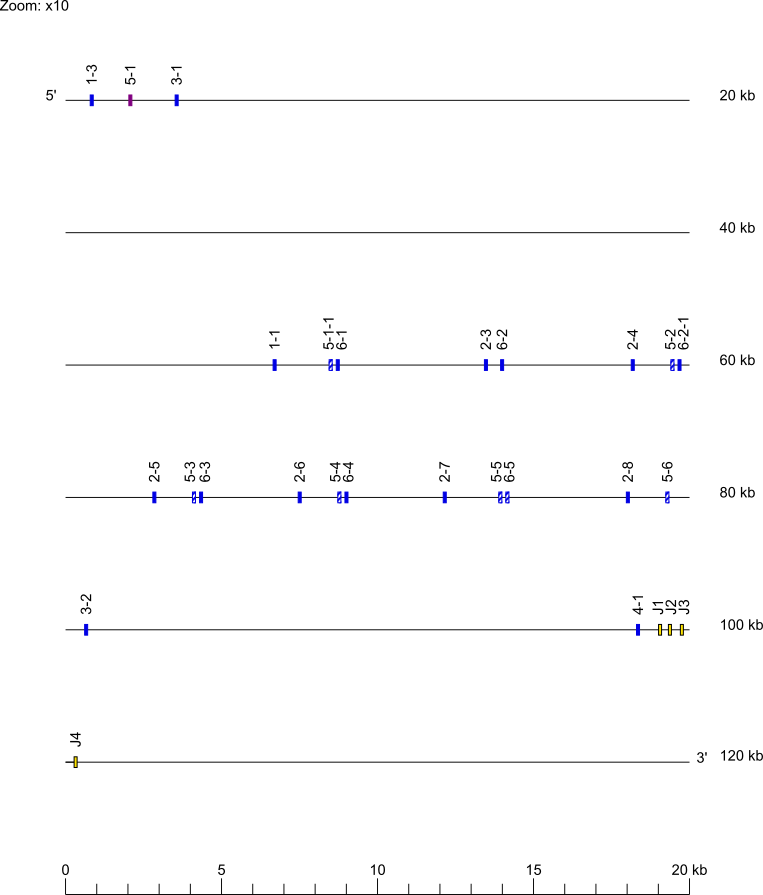
Zoom for IGHD and IGHJ genes
IGHV genes
- [1] Crews, S. et al., Cell, 25, 59-66 (1981).
- [2] Brodeur, P.H. and Riblet, R., Eur. J. Immunol. 14, 922-930 (1984).
- [3] Winter, E. et al., EMBO J., 4, 2861-2867 (1985).
- [4] Brodeur, P.H. et al., J. Exp. Med., 168, 2261-2278 (1988).
- [5] Kofler, R. et al., J. Immunol., 140, 4031-4034 (1988).
- [6] Pennell, C.A. et al., Eur. J. Immunol., 19, 2115-2121 (1989).
- [7] Tutter, A. et al., J. Immunol., 147, 3215-3223 (1991).
- [8] Kofler, R. et al., Immunol. Rev., 128, 5-21 (1992).
- [9] Sims, M.J. et al., J. Immunol., 149, 1642-1648 (1992).
- [10] Sheehan, K.M. et al., J. Immunol., 151, 5364-5375 (1993).
- [11] Mainville, C.A. et al., J. Immunol., 156, 1058-1046 (1996).
- [12] Almagro, J.C. et al., Mol. Immunol., 34, 119-1214 (1997).
- [13] Kurosawa, Y. and Tonegawa, S., J. Exp. Med., 155, 201-218 (1982).
- [14] Woods, C. and Tonegawa, S., Proc. Natl. Acad. Sci. USA 80, 3030-3034 (1983).
- [15] Feeney, A.J and Ribbet, R., Immunogenetics, 37, 217-221 (1993).
- [16] Early, P. et al., Cell, 19, 981-992 (1980).
- [17] Newell, N. et al., Science, 209, 1128-1132 (1980).
- [18] Sakano, H. et al., Nature, 286, 676-683 (1980).
- [19] Edelman, G.M. et al., Proc. Natl. Acad. Sci. USA, 63, 78-85 (1969).
- [20] Honjo, T. and Kataoka,T., Proc. Natl Acad. Sci. USA, 75, 2140-2144 (1978).
- [21] Honjo, T. et al., Immunological Rev., 59, 33-67 (1981).
- [22] Yamawaki-Kataoka, Y. et al., Proc. Natl. Acad. Sci. USA, 79, 2623-2627 (1982).
- [23] Shimiza, A. et al., Cell, 28, 499-506 (1982).
- [24] Riblet, R. In: Molecular Biology of B cells. Honjo, T., Alt, F.W. and Neuberger, M. eds, Elsevier Academic Press, London, pp 19-26 (2004).
- [25] Folch, G., Giudicelli, V., Jean, C., Ginestoux, C., Lefranc G. and Lefranc, M.-P. IMGT overview: the mouse immunoglobulin heavy IGH genes.19th International Mouse Genome Conference (IMGC), November 5-8, 2005, Strasbourg, France (2005).
 .
.
- Locus representation: Mouse (Mus musculus) IGH anterior to November 2005.
- Gene table: House mouse (Mus musculus) IGHV
- Gene table: House mouse (Mus musculus) IGHD
- Gene table: House mouse (Mus musculus) IGHJ
- Gene table: House mouse (Mus musculus) IGHC
- Potential germline repertoire: House mouse (Mus musculus) IGHV, IGHD and IGHJ genes on chromosome 12
- Created:
- 05/11/2005
- Last updated:
- 22/09/2023
- Authors:
- Géraldine Folch
